Alterations of the Gut Microbiota in Patients with Diabetic Nephropathy
- PMID: 35863004
- PMCID: PMC9430528
- DOI: 10.1128/spectrum.00324-22
Alterations of the Gut Microbiota in Patients with Diabetic Nephropathy
Abstract
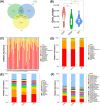
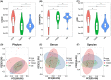
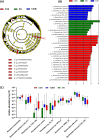
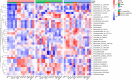
Diabetic nephropathy (DN) is the primary cause of end-stage renal disease. Accumulating studies have implied a critical role for the gut microbiota in diabetes mellitus (DM) and DN. However, the precise roles and regulatory mechanisms of the gut microbiota in the pathogenesis of DN remain largely unclear. In this study, metagenomics sequencing was performed using fecal samples from healthy controls (CON) and type 2 diabetes mellitus (T2DM) patients with or without DN. Fresh fecal samples from 15 T2DM patients without DN, 15 DN patients, and 15 age-, gender-, and body mass index (BMI)-matched healthy controls were collected. The compositions and potential functions of the gut microbiota were estimated. Although no difference of gut microbiota α and β diversity was observed between the CON, T2DM, and DN groups, the relative abundances of butyrate-producing bacteria (Clostridium, Eubacterium, and Roseburia intestinalis) and potential probiotics (Lachnospira and Intestinibacter) were significantly reduced in T2DM and DN patients. Besides, Bacteroides stercoris was significantly enriched in fecal samples from patients with DN. Moreover, Clostridium sp. 26_22 was negatively associated with serum creatinine (P < 0.05). DN patients could be accurately distinguished from CON by Clostridium sp. CAG_768 (area under the curve [AUC] = 0.941), Bacteroides propionicifaciens (AUC = 0.905), and Clostridium sp. CAG_715 (AUC = 0.908). DN patients could be accurately distinguished from T2DM patients by Pseudomonadales, Fusobacterium varium, and Prevotella sp. MSX73 (AUC = 0.889). Regarding the potential bacterial functions of the gut microbiota, the citrate cycle, base excision repair, histidine metabolism, lipoic acid metabolism, and bile acid biosynthesis were enriched in DN patients, while selenium metabolism and branched-chain amino acid biosynthesis were decreased in DN patients. IMPORTANCE Gut microbiota imbalance is found in fecal samples from DN patients, in which Roseburia intestinalis is significantly decreased, while Bacteroides stercoris is increased. There is a significant correlation between gut microbiota imbalance and clinical indexes related to lipid metabolism, glucose metabolism, and renal function. The gut microbiota may be predictive factors for the development and progression of DN, although further studies are warranted to illustrate their regulatory mechanisms.
Keywords: composition; diabetic nephropathy; function; gut microbiota; metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition.Acta Diabetol. 2019 May;56(5):581-592. doi: 10.1007/s00592-019-01316-7. Epub 2019 Mar 19. Acta Diabetol. 2019. PMID: 30888537
-
Alterations of gut microbiota in biopsy-proven diabetic nephropathy and a long history of diabetes without kidney damage.Sci Rep. 2023 Jul 27;13(1):12150. doi: 10.1038/s41598-023-39444-4. Sci Rep. 2023. PMID: 37500743 Free PMC article.
-
Characteristics of Gut Microbiota in Female Patients with Diabetic Microvascular Complications.J Diabetes Res. 2022 Oct 27;2022:2980228. doi: 10.1155/2022/2980228. eCollection 2022. J Diabetes Res. 2022. PMID: 36339086 Free PMC article.
-
Gut microbiota microbial metabolites in diabetic nephropathy patients: far to go.Front Cell Infect Microbiol. 2024 May 8;14:1359432. doi: 10.3389/fcimb.2024.1359432. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38779567 Free PMC article. Review.
-
Effect of traditional Chinese medicine on gut microbiota in adults with type 2 diabetes: A systematic review and meta-analysis.Phytomedicine. 2021 Jul 15;88:153455. doi: 10.1016/j.phymed.2020.153455. Epub 2020 Dec 30. Phytomedicine. 2021. PMID: 33478831
Cited by
-
The Causal Relationship Between Skin Microbiota and Facial Aging: A Mendelian Randomization Study.Aesthetic Plast Surg. 2024 Dec;48(24):5350-5357. doi: 10.1007/s00266-024-04217-5. Epub 2024 Jul 8. Aesthetic Plast Surg. 2024. PMID: 38977452
-
Genetic Evidence for the Causal Relationship Between Gut Microbiota and Diabetic Kidney Disease: A Bidirectional, Two-Sample Mendelian Randomisation Study.J Diabetes Res. 2024 Oct 23;2024:4545595. doi: 10.1155/2024/4545595. eCollection 2024. J Diabetes Res. 2024. PMID: 39479291 Free PMC article.
-
Advances in fecal microbiota transplantation for the treatment of diabetes mellitus.Front Cell Infect Microbiol. 2024 Apr 10;14:1370999. doi: 10.3389/fcimb.2024.1370999. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38660489 Free PMC article. Review.
-
Astragalus Mongholicus Polysaccharides Alleviate Kidney Injury in Rats with Type 2 Diabetes Through Modulation of Oxidation, Inflammation, and Gut Microbiota.Int J Mol Sci. 2025 Feb 10;26(4):1470. doi: 10.3390/ijms26041470. Int J Mol Sci. 2025. PMID: 40003935 Free PMC article.
-
Exploring the beneficial effect of gut microbiota metabolites on diabetic nephropathy via network pharmacology study.Sci Rep. 2025 Apr 1;15(1):11027. doi: 10.1038/s41598-025-95824-y. Sci Rep. 2025. PMID: 40164705 Free PMC article.
References
-
- Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, Colagiuri S, Guariguata L, Motala AA, Ogurtsova K, Shaw JE, Bright D, Williams R, IDF Diabetes Atlas Committee. . 2019. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract 157:107843. doi:10.1016/j.diabres.2019.107843. - DOI - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

