Rapid Detection of the Omicron (B.1.1.529) SARS-CoV-2 Variant Using a COVID-19 Diagnostic PCR Assay
- PMID: 35863025
- PMCID: PMC9430347
- DOI: 10.1128/spectrum.00990-22
Rapid Detection of the Omicron (B.1.1.529) SARS-CoV-2 Variant Using a COVID-19 Diagnostic PCR Assay
Abstract
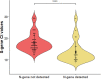
The Omicron (B.1.1.529) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the last variant of concern (VOC) identified to date. Compared to whole-genome or gene-specific sequencing methods, reverse-transcription PCR assays may be a simpler approach to study VOCs. We used a point-of-care COVID-19 diagnostic PCR assay to detect the Omicron SARS-CoV-2 variant in the respiratory tract samples of COVID-19 patients who had tested positive for SARS-CoV-2 RNA between April 2021 and January 2022. Sequencing analyses had shown that 87 samples were positive for the Omicron variant and 43 samples were positive for a non-Omicron variant (Delta, 18 samples; Alpha, 13 samples; Gamma, 10 samples; Beta, 1 sample; or Epsilon, 1 sample). According to results by the PCR assay, whose primers anneal a nucleocapsid (N) gene region that comprises the E31/R32/S33 deletion (also termed the del31/33 mutation), we found that N gene target failure/dropout (i.e., a negative/low result) occurred in 86 (98.8%) of 87 Omicron variant-positive samples tested. These results were assessed in relation to those of the spike (S) gene, which expectedly, was detected in all (100%) 130 samples. A total of 43 (100%) of 43 Delta, Alpha, Gamma, Beta, or Epsilon variant-positive samples had a positive result with the N gene. Importantly, in 86 of 87 Omicron variant-positive samples, the del31/33 mutation was detected together with a P13L mutation, which was, instead, detected alone in the Omicron variant-positive sample that had a positive N-gene result. IMPORTANCE Rapid detection of the Omicron SARS-CoV-2 variant in patients' respiratory tract samples may influence therapeutic choices, because this variant is known to escape from certain monoclonal antibodies. Our findings strengthen the importance of manufacturers' efforts to improve the existing COVID-19 diagnostic PCR assays and/or to develop novel variant-specific PCR assays. Furthermore, our findings show that only a small fraction of SARS-CoV-2-positive samples may require whole-genome sequencing analysis, which is still crucial to validate PCR assay results. We acknowledge that the emergence of novel variants containing mutations outside the PCR assay target region could, however, allow an assay to work as per specifications without being able to identify a SARS-CoV-2-positive sample as a variant. Future work and more experience in this topic will help to reduce the risk of misidentification of SARS-CoV-2 variants that is unavoidable when using the current PCR assays.
Keywords: Omicron SARS-CoV-2 variant; PCR assay; rapid testing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Two-Period Study Results from a Large Italian Hospital Laboratory Attesting SARS-CoV-2 Variant PCR Assay Evolution.Microbiol Spectr. 2022 Dec 21;10(6):e0292222. doi: 10.1128/spectrum.02922-22. Epub 2022 Nov 21. Microbiol Spectr. 2022. PMID: 36409091 Free PMC article.
-
Accessible and Adaptable Multiplexed Real-Time PCR Approaches to Identify SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2022 Oct 26;10(5):e0322222. doi: 10.1128/spectrum.03222-22. Epub 2022 Sep 15. Microbiol Spectr. 2022. PMID: 36106882 Free PMC article.
-
In silico evaluation of the impact of Omicron variant of concern sublineage BA.4 and BA.5 on the sensitivity of RT-qPCR assays for SARS-CoV-2 detection using whole genome sequencing.J Med Virol. 2023 Jan;95(1):e28241. doi: 10.1002/jmv.28241. Epub 2022 Nov 8. J Med Virol. 2023. PMID: 36263448 Free PMC article.
-
Structural impact of a new spike Y170W mutation detected in early emerging SARS-CoV-2 Omicron variants in France.Virus Res. 2024 May;343:199354. doi: 10.1016/j.virusres.2024.199354. Epub 2024 Mar 16. Virus Res. 2024. PMID: 38492859 Free PMC article. Review.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
Cited by
-
Systematic Exploration of SARS-CoV-2 Adaptation to Vero E6, Vero E6/TMPRSS2, and Calu-3 Cells.Genome Biol Evol. 2023 Apr 6;15(4):evad035. doi: 10.1093/gbe/evad035. Genome Biol Evol. 2023. PMID: 36852863 Free PMC article.
-
Efficient Tracing of the SARS-CoV-2 Omicron Variants in Santa Barbara County Using a Rapid Quantitative Reverse Transcription PCR Assay.Diagnostics (Basel). 2022 Nov 15;12(11):2805. doi: 10.3390/diagnostics12112805. Diagnostics (Basel). 2022. PMID: 36428863 Free PMC article.
-
Mutational Analysis of Circulating Omicron SARS-CoV-2 Lineages in the Al-Baha Region of Saudi Arabia.J Multidiscip Healthc. 2023 Jul 27;16:2117-2136. doi: 10.2147/JMDH.S419859. eCollection 2023. J Multidiscip Healthc. 2023. PMID: 37529147 Free PMC article.
-
A review of current effective COVID-19 testing methods and quality control.Arch Microbiol. 2023 May 17;205(6):239. doi: 10.1007/s00203-023-03579-9. Arch Microbiol. 2023. PMID: 37195393 Free PMC article. Review.
-
Sensitive detection of SARS-CoV-2 main protease 3CLpro with an engineered ribonuclease zymogen.Protein Sci. 2024 Apr;33(4):e4916. doi: 10.1002/pro.4916. Protein Sci. 2024. PMID: 38501598 Free PMC article.
References
-
- World Health Organization. 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1..... Accessed 8 March 2022.
-
- CoVariants. 2022. Overview of variants/mutations. https://covariants.org/variants. Accessed 25 May 2022.
-
- U.S. Food and Drug Administration. 2022. Coronavirus (COVID-19) update: FDA limits use of certain monoclonal antibodies to treat COVID-19 due to the Omicron variant. www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-.... Accessed 8 March 2022.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

