G protein-coupled receptor interactions with arrestins and GPCR kinases: The unresolved issue of signal bias
- PMID: 35863432
- PMCID: PMC9418498
- DOI: 10.1016/j.jbc.2022.102279
G protein-coupled receptor interactions with arrestins and GPCR kinases: The unresolved issue of signal bias
Abstract
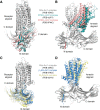
G protein-coupled receptor (GPCR) kinases (GRKs) and arrestins interact with agonist-bound GPCRs to promote receptor desensitization and downregulation. They also trigger signaling cascades distinct from those of heterotrimeric G proteins. Biased agonists for GPCRs that favor either heterotrimeric G protein or GRK/arrestin signaling are of profound pharmacological interest because they could usher in a new generation of drugs with greatly reduced side effects. One mechanism by which biased agonism might occur is by stabilizing receptor conformations that preferentially bind to GRKs and/or arrestins. In this review, we explore this idea by comparing structures of GPCRs bound to heterotrimeric G proteins with those of the same GPCRs in complex with arrestins and GRKs. The arrestin and GRK complexes all exhibit high conformational heterogeneity, which is likely a consequence of their unusual ability to adapt and bind to hundreds of different GPCRs. This dynamic behavior, along with the experimental tactics required to stabilize GPCR complexes for biophysical analysis, confounds these comparisons, but some possible molecular mechanisms of bias are beginning to emerge. We also examine if and how the recent structures advance our understanding of how arrestins parse the "phosphorylation barcodes" installed in the intracellular loops and tails of GPCRs by GRKs. In the future, structural analyses of arrestins in complex with intact receptors that have well-defined native phosphorylation barcodes, such as those installed by the two nonvisual subfamilies of GRKs, will be particularly illuminating.
Keywords: G protein-coupled receptor; GPCR kinase; GRK; allostery; arrestin; biased agonism; cryo-electron microscopy; desensitization; single particle reconstruction.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



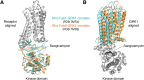

Similar articles
-
How Ligands Achieve Biased Signaling toward Arrestins.Biochemistry. 2025 Mar 4;64(5):967-977. doi: 10.1021/acs.biochem.4c00843. Epub 2025 Feb 12. Biochemistry. 2025. PMID: 39943784 Review.
-
G protein-coupled receptor kinases (GRKs) orchestrate biased agonism at the β2-adrenergic receptor.Sci Signal. 2018 Aug 21;11(544):eaar7084. doi: 10.1126/scisignal.aar7084. Sci Signal. 2018. PMID: 30131371
-
Phosphorylation barcoding as a mechanism of directing GPCR signaling.Sci Signal. 2011 Aug 9;4(185):pe36. doi: 10.1126/scisignal.2002331. Sci Signal. 2011. PMID: 21868354 Review.
-
Feedback regulation of G protein-coupled receptor signaling by GRKs and arrestins.Semin Cell Dev Biol. 2016 Feb;50:95-104. doi: 10.1016/j.semcdb.2015.12.015. Epub 2016 Jan 7. Semin Cell Dev Biol. 2016. PMID: 26773211 Free PMC article. Review.
-
GRKs as Modulators of Neurotransmitter Receptors.Cells. 2020 Dec 31;10(1):52. doi: 10.3390/cells10010052. Cells. 2020. PMID: 33396400 Free PMC article. Review.
Cited by
-
ACKR3-arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding.bioRxiv [Preprint]. 2023 Jul 19:2023.07.18.549504. doi: 10.1101/2023.07.18.549504. bioRxiv. 2023. PMID: 37502840 Free PMC article. Preprint.
-
Investigation of adenosine A1 receptor-mediated β-arrestin 2 recruitment using a split-luciferase assay.Front Pharmacol. 2023 May 30;14:1172551. doi: 10.3389/fphar.2023.1172551. eCollection 2023. Front Pharmacol. 2023. PMID: 37324481 Free PMC article.
-
Molecular basis of β-arrestin coupling to the metabotropic glutamate receptor mGlu3.Nat Chem Biol. 2025 Aug;21(8):1262-1269. doi: 10.1038/s41589-025-01858-8. Epub 2025 Mar 6. Nat Chem Biol. 2025. PMID: 40050438
-
Computational drug development for membrane protein targets.Nat Biotechnol. 2024 Feb;42(2):229-242. doi: 10.1038/s41587-023-01987-2. Epub 2024 Feb 15. Nat Biotechnol. 2024. PMID: 38361054 Review.
-
Know your molecule: pharmacological characterization of drug candidates to enhance efficacy and reduce late-stage attrition.Nat Rev Drug Discov. 2024 Aug;23(8):626-644. doi: 10.1038/s41573-024-00958-9. Epub 2024 Jun 18. Nat Rev Drug Discov. 2024. PMID: 38890494 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

