Three-Dimensional Visualization of Viral Structure, Entry, and Replication Underlying the Spread of SARS-CoV-2
- PMID: 35863749
- PMCID: PMC9344915
- DOI: 10.1021/acs.chemrev.1c01062
Three-Dimensional Visualization of Viral Structure, Entry, and Replication Underlying the Spread of SARS-CoV-2
Abstract
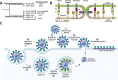
The global spread of SARS-CoV-2 has proceeded at an unprecedented rate. Remarkably, characterization of the virus using modern tools in structural biology has also progressed at exceptional speed. Advances in electron-based imaging techniques, combined with decades of foundational studies on related viruses, have enabled the research community to rapidly investigate structural aspects of the novel coronavirus from the level of individual viral proteins to imaging the whole virus in a native context. Here, we provide a detailed review of the structural biology and pathobiology of SARS-CoV-2 as it relates to all facets of the viral life cycle, including cell entry, replication, and three-dimensional (3D) packaging based on insights obtained from X-ray crystallography, cryo-electron tomography, and single-particle cryo-electron microscopy. The structural comparison between SARS-CoV-2 and the related earlier viruses SARS-CoV and MERS-CoV is a common thread throughout this review. We conclude by highlighting some of the outstanding unanswered structural questions and underscore areas that are under rapid current development such as the design of effective therapeutics that block viral infection.
Conflict of interest statement
The authors declare the following competing financial interest(s): S.S. is the founder and CEO of Gandeeva Therapeutics Inc.
Figures




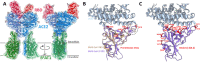



Similar articles
-
Structural biology of SARS-CoV-2.Prog Mol Biol Transl Sci. 2024;202:31-43. doi: 10.1016/bs.pmbts.2023.11.001. Epub 2024 Jan 3. Prog Mol Biol Transl Sci. 2024. PMID: 38237989 Review.
-
Structural insights into SARS-CoV-2 proteins.J Mol Biol. 2021 Jan 22;433(2):166725. doi: 10.1016/j.jmb.2020.11.024. Epub 2020 Nov 24. J Mol Biol. 2021. PMID: 33245961 Free PMC article. Review.
-
A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites.Immunobiology. 2023 Jan;228(1):152302. doi: 10.1016/j.imbio.2022.152302. Epub 2022 Nov 15. Immunobiology. 2023. PMID: 36434912 Free PMC article. Review.
-
A structural view of the SARS-CoV-2 virus and its assembly.Curr Opin Virol. 2022 Feb;52:123-134. doi: 10.1016/j.coviro.2021.11.011. Epub 2021 Dec 4. Curr Opin Virol. 2022. PMID: 34915287 Free PMC article. Review.
-
Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress.Nat Commun. 2021 Jul 30;12(1):4629. doi: 10.1038/s41467-021-24887-y. Nat Commun. 2021. PMID: 34330917 Free PMC article.
Cited by
-
Natural and Synthetic Coumarins as Potential Drug Candidates against SARS-CoV-2/COVID-19.Curr Med Chem. 2025;32(3):539-562. doi: 10.2174/0109298673285609231220111556. Curr Med Chem. 2025. PMID: 38243979 Review.
-
COVID-19 Biogenesis and Intracellular Transport.Int J Mol Sci. 2023 Feb 24;24(5):4523. doi: 10.3390/ijms24054523. Int J Mol Sci. 2023. PMID: 36901955 Free PMC article. Review.
-
Electron Tomography as a Tool to Study SARS-CoV-2 Morphology.Int J Mol Sci. 2024 Nov 1;25(21):11762. doi: 10.3390/ijms252111762. Int J Mol Sci. 2024. PMID: 39519314 Free PMC article. Review.
-
Multidimensional futuristic approaches to address the pandemics beyond COVID-19.Heliyon. 2023 Jun;9(6):e17148. doi: 10.1016/j.heliyon.2023.e17148. Epub 2023 Jun 11. Heliyon. 2023. PMID: 37325452 Free PMC article. Review.
-
SARS-CoV-2 epitope-specific T cells: Immunity response feature, TCR repertoire characteristics and cross-reactivity.Front Immunol. 2023 Mar 10;14:1146196. doi: 10.3389/fimmu.2023.1146196. eCollection 2023. Front Immunol. 2023. PMID: 36969254 Free PMC article. Review.
References
-
- Zhong N.; Zheng B.; Li Y.; Poon L.; Xie Z.; Chan K.; Li P.; Tan S.; Chang Q.; Xie J.; et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003, 362, 1353–1358. 10.1016/S0140-6736(03)14630-2. - DOI - PMC - PubMed
-
- Listings of WHO’s Response to COVID-19. World Health Organization, June 29, 2020. https://www.who.int/news/item/29-06-2020-covidtimeline (accessed 2021-09-07).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

