Therapeutic targeting of PD-1/PD-L1 blockade by novel small-molecule inhibitors recruits cytotoxic T cells into solid tumor microenvironment
- PMID: 35863821
- PMCID: PMC9310269
- DOI: 10.1136/jitc-2022-004695
Therapeutic targeting of PD-1/PD-L1 blockade by novel small-molecule inhibitors recruits cytotoxic T cells into solid tumor microenvironment
Abstract
Background: Inhibiting programmed cell death protein 1 (PD-1) or PD-ligand 1 (PD-L1) has shown exciting clinical outcomes in diverse human cancers. So far, only monoclonal antibodies are approved as PD-1/PD-L1 inhibitors. While significant clinical outcomes are observed on patients who respond to these therapeutics, a large proportion of the patients do not benefit from the currently available immune checkpoint inhibitors, which strongly emphasize the importance of developing new immunotherapeutic agents.
Methods: In this study, we followed a transdisciplinary approach to discover novel small molecules that can modulate PD-1/PD-L1 interaction. To that end, we employed in silico analyses combined with in vitro, ex vivo, and in vivo experimental studies to assess the ability of novel compounds to modulate PD-1/PD-L1 interaction and enhance T-cell function.
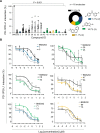
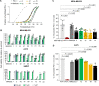
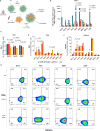
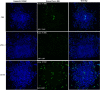
Results: Accordingly, in this study we report the identification of novel small molecules, which like anti-PD-L1/PD-1 antibodies, can stimulate human adaptive immune responses. Unlike these biological compounds, our newly-identified small molecules enabled an extensive infiltration of T lymphocytes into three-dimensional solid tumor models, and the recruitment of cytotoxic T lymphocytes to the tumor microenvironment in vivo, unveiling a unique potential to transform cancer immunotherapy.
Conclusions: We identified a new promising family of small-molecule candidates that regulate the PD-L1/PD-1 signaling pathway, promoting an extensive infiltration of effector CD8 T cells to the tumor microenvironment.
Keywords: IMMUNOLOGY; Lymphocyte Activation; Lymphocytes, Tumor-Infiltrating; Programmed Cell Death 1 Receptor; Tumor Escape.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: RS-F is a Board Director at Teva Pharmaceutical Industries. All other authors declare that they have no competing interests.
Figures


Similar articles
-
Uncovering the colorectal cancer immunotherapeutic potential: Evening primrose (Oenothera biennis) root extract and its active compound oenothein B targeting the PD-1/PD-L1 blockade.Phytomedicine. 2024 Mar;125:155370. doi: 10.1016/j.phymed.2024.155370. Epub 2024 Jan 17. Phytomedicine. 2024. PMID: 38266440
-
Roles of PD-1/PD-L1 Pathway: Signaling, Cancer, and Beyond.Adv Exp Med Biol. 2020;1248:33-59. doi: 10.1007/978-981-15-3266-5_3. Adv Exp Med Biol. 2020. PMID: 32185706 Review.
-
Inhibition of T-cell-mediated immune response via the PD-1/ PD-L1 axis in cholangiocarcinoma cells.Eur J Pharmacol. 2021 Apr 15;897:173960. doi: 10.1016/j.ejphar.2021.173960. Epub 2021 Feb 19. Eur J Pharmacol. 2021. PMID: 33617828
-
Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma.J Hepatol. 2019 Aug;71(2):333-343. doi: 10.1016/j.jhep.2019.04.007. Epub 2019 May 7. J Hepatol. 2019. PMID: 31071366
-
The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma.Clin Ther. 2015 Apr 1;37(4):764-82. doi: 10.1016/j.clinthera.2015.02.018. Epub 2015 Mar 29. Clin Ther. 2015. PMID: 25823918 Free PMC article. Review.
Cited by
-
Enhancement of Immunotherapies in Head and Neck Cancers Using Biomaterial-Based Treatment Strategies.Tissue Eng Part C Methods. 2023 Jun;29(6):257-275. doi: 10.1089/ten.TEC.2023.0090. Tissue Eng Part C Methods. 2023. PMID: 37183412 Free PMC article. Review.
-
Computational identification of PDL1 inhibitors and their cytotoxic effects with silver and gold nanoparticles.Sci Rep. 2024 Nov 4;14(1):26610. doi: 10.1038/s41598-024-77868-8. Sci Rep. 2024. PMID: 39496756 Free PMC article.
-
Theoretical and experimental studies on the interaction of biphenyl ligands with human and murine PD-L1: Up-to-date clues for drug design.Comput Struct Biotechnol J. 2023 Jun 8;21:3355-3368. doi: 10.1016/j.csbj.2023.06.006. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37384351 Free PMC article.
-
Lipid metabolic features of T cells in the Tumor Microenvironment.Lipids Health Dis. 2022 Oct 6;21(1):94. doi: 10.1186/s12944-022-01705-y. Lipids Health Dis. 2022. PMID: 36203151 Free PMC article. Review.
-
Effectiveness and safety of PD-1/PD-L1 inhibitors in advanced or recurrent endometrial cancer: a systematic review and meta-analysis.Front Pharmacol. 2023 Dec 14;14:1330877. doi: 10.3389/fphar.2023.1330877. eCollection 2023. Front Pharmacol. 2023. PMID: 38161705 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
