Heterogeneous nuclear ribonucleoprotein U (HNRNPU) safeguards the developing mouse cortex
- PMID: 35864088
- PMCID: PMC9304408
- DOI: 10.1038/s41467-022-31752-z
Heterogeneous nuclear ribonucleoprotein U (HNRNPU) safeguards the developing mouse cortex
Abstract
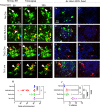
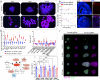
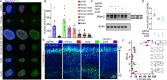
HNRNPU encodes the heterogeneous nuclear ribonucleoprotein U, which participates in RNA splicing and chromatin organization. Microdeletions in the 1q44 locus encompassing HNRNPU and other genes and point mutations in HNRNPU cause brain disorders, including early-onset seizures and severe intellectual disability. We aimed to understand HNRNPU's roles in the developing brain. Our work revealed that HNRNPU loss of function leads to rapid cell death of both postmitotic neurons and neural progenitors, with an apparent higher sensitivity of the latter. Further, expression and alternative splicing of multiple genes involved in cell survival, cell motility, and synapse formation are affected following Hnrnpu's conditional truncation. Finally, we identified pharmaceutical and genetic agents that can partially reverse the loss of cortical structures in Hnrnpu mutated embryonic brains, ameliorate radial neuronal migration defects and rescue cultured neural progenitors' cell death.
© 2022. The Author(s).
Conflict of interest statement
D.B.G. is the founder of Praxis Precision Medicines and Action Biosciences. O.D. receives grant support from NINDS, NIMH, MURI, CDC, and NSF. He has equity and/or compensation from the following companies: Tilray, Receptor Life Sciences, Qstate Biosciences, Hitch Biosciences, Tevard Biosciences, Regel Biosciences, Script Biosciences, Actio Biosciences, Empatica, SilverSpike, and California Cannabis Enterprises (CCE). He has received consulting fees from Zogenix, Ultragenyx, BridgeBio, and Marinus. He holds patents for cannabidiol in treating neurological disorders, but these are owned by GW Pharmaceuticals, and he has waived any financial interests. He holds other patents in molecular biology. He is the managing partner of the PhiFund Ventures. Ingrid Scheffer has served on scientific advisory boards for BioMarin, Chiesi, Eisai, Encoded Therapeutics, GlaxoSmithKline, Knopp Biosciences, Nutricia, Rogcon, Takeda Pharmaceuticals, UCB, Xenon Pharmaceuticals; has received speaker honoraria from GlaxoSmithKline, UCB, BioMarin, Biocodex, Chiesi, Liva Nova and Eisai; has received funding for travel from UCB, Biocodex, GlaxoSmithKline, Biomarin and Eisai; has served as an investigator for Anavex Life Sciences, Cerecin Inc, Cereval Therapeutics, Eisai, Encoded Therapeutics, EpiMinder Inc, Epygenyx, ES-Therapeutics, GW Pharma, Marinus, Neurocrine BioSciences, Ovid Therapeutics, Takeda Pharmaceuticals, UCB, Ultragenyx, Xenon Pharmaceutical, Zogenix and Zynerba; and has consulted for Atheneum Partners, Care Beyond Diagnosis, Epilepsy Consortium, Ovid Therapeutics, UCB and Zynerba Pharmaceuticals; and is a Non-Executive Director of Bellberry Ltd and a Director of the Australian Academy of Health and Medical Sciences and the Australian Council of Learned Academies Limited. The other authors declare no competing interests.
Figures



Similar articles
-
HNRNPU's multi-tasking is essential for proper cortical development.Bioessays. 2023 Sep;45(9):e2300039. doi: 10.1002/bies.202300039. Epub 2023 Jul 13. Bioessays. 2023. PMID: 37439444
-
Heterozygous HNRNPU variants cause early onset epilepsy and severe intellectual disability.Hum Genet. 2017 Jul;136(7):821-834. doi: 10.1007/s00439-017-1795-6. Epub 2017 Apr 9. Hum Genet. 2017. PMID: 28393272
-
Deficiency of the Heterogeneous Nuclear Ribonucleoprotein U locus leads to delayed hindbrain neurogenesis.Biol Open. 2023 Oct 15;12(10):bio060113. doi: 10.1242/bio.060113. Epub 2023 Oct 10. Biol Open. 2023. PMID: 37815090 Free PMC article.
-
Expanding the Phenotypic Spectrum of HNRNPU-Related Disorder, Documenting the First Familial Presentation and Comprehensive Review.Am J Med Genet A. 2025 Jun;197(6):e64013. doi: 10.1002/ajmg.a.64013. Epub 2025 Feb 20. Am J Med Genet A. 2025. PMID: 39976380 Review.
-
Clinical findings of 21 previously unreported probands with HNRNPU-related syndrome and comprehensive literature review.Am J Med Genet A. 2020 Jul;182(7):1637-1654. doi: 10.1002/ajmg.a.61599. Epub 2020 Apr 22. Am J Med Genet A. 2020. PMID: 32319732 Review.
Cited by
-
Identification of MicroRNA Profiles in Fetal Spina Bifida: The Role in Pathomechanism and Diagnostic Significance.Int J Mol Sci. 2024 Mar 1;25(5):2896. doi: 10.3390/ijms25052896. Int J Mol Sci. 2024. PMID: 38474143 Free PMC article.
-
Germline pathogenic variants in HNRNPU are associated with alterations in blood methylome.Eur J Hum Genet. 2023 Sep;31(9):1040-1047. doi: 10.1038/s41431-023-01422-9. Epub 2023 Jul 5. Eur J Hum Genet. 2023. PMID: 37407733 Free PMC article.
-
Role of the SAF-A SAP domain in X inactivation, transcription, splicing, and cell proliferation.bioRxiv [Preprint]. 2024 Sep 10:2024.09.09.612041. doi: 10.1101/2024.09.09.612041. bioRxiv. 2024. Update in: PLoS Genet. 2025 Jun 10;21(6):e1011719. doi: 10.1371/journal.pgen.1011719. PMID: 39314300 Free PMC article. Updated. Preprint.
-
Neurodevelopmental deficits and cell-type-specific transcriptomic perturbations in a mouse model of HNRNPU haploinsufficiency.PLoS Genet. 2023 Oct 2;19(10):e1010952. doi: 10.1371/journal.pgen.1010952. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37782669 Free PMC article.
-
Comprehensive transcriptome analysis reveals altered mRNA splicing and post-transcriptional changes in the aged mouse brain.Nucleic Acids Res. 2024 Apr 12;52(6):2865-2885. doi: 10.1093/nar/gkae172. Nucleic Acids Res. 2024. PMID: 38471806 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

