A metagenomic DNA sequencing assay that is robust against environmental DNA contamination
- PMID: 35864089
- PMCID: PMC9304412
- DOI: 10.1038/s41467-022-31654-0
A metagenomic DNA sequencing assay that is robust against environmental DNA contamination
Abstract
Metagenomic DNA sequencing is a powerful tool to characterize microbial communities but is sensitive to environmental DNA contamination, in particular when applied to samples with low microbial biomass. Here, we present Sample-Intrinsic microbial DNA Found by Tagging and sequencing (SIFT-seq) a metagenomic sequencing assay that is robust against environmental DNA contamination introduced during sample preparation. The core idea of SIFT-seq is to tag the DNA in the sample prior to DNA isolation and library preparation with a label that can be recorded by DNA sequencing. Any contaminating DNA that is introduced in the sample after tagging can then be bioinformatically identified and removed. We applied SIFT-seq to screen for infections from microorganisms with low burden in blood and urine, to identify COVID-19 co-infection, to characterize the urinary microbiome, and to identify microbial DNA signatures of sepsis and inflammatory bowel disease in blood.
© 2022. The Author(s).
Conflict of interest statement
I.D.V., O.M., A.P.C., and A.C. have submitted a patent related to the present work. A.P.C., I.D.V., D.D., and J.R.L. are inventors on the patent US-2020-0048713-A1 titled “Methods of Detecting Cell-Free DNA in Biological Samples.” I.D.V. is a member of the Scientific Advisory Board of Karius Inc., Kanvas Biosciences and GenDX. J.R.L. received research support under an investigator-initiated research grant from BioFire Diagnostics, LLC. E.J.S. is a consultant for Axle Informatics. Remaining authors declare no competing interests.
Figures
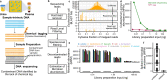
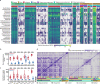


Update of
-
A metagenomic DNA sequencing assay that is robust against environmental DNA contamination.bioRxiv [Preprint]. 2021 Nov 23:2021.11.22.469599. doi: 10.1101/2021.11.22.469599. bioRxiv. 2021. Update in: Nat Commun. 2022 Jul 21;13(1):4197. doi: 10.1038/s41467-022-31654-0. PMID: 34845444 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

