Improving genetic diagnosis by disease-specific, ACMG/AMP variant interpretation guidelines for hearing loss
- PMID: 35864128
- PMCID: PMC9304357
- DOI: 10.1038/s41598-022-16661-x
Improving genetic diagnosis by disease-specific, ACMG/AMP variant interpretation guidelines for hearing loss
Abstract
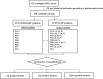
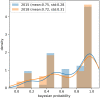
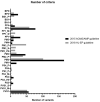
The 2018 Hearing Loss Expert Panel (HL-EP)-specific guidelines specified from the universal 2015 ACMG/AMP guidelines are proposed to be used in genetic HL, which prompted this study. A genetic HL cohort comprising 135 unrelated probands with available exome sequencing data was established. Overall, 169 variants were prioritized as candidates and interpreted using the 2015 ACMG/AMP and 2018 HL-EP guidelines. Changes in rule application and variant classification between the guidelines were compared. The concordance rate of variant classification of each variant between the guidelines was 71.60%, with significant difference. The proportion of pathogenic variants increased from 13.02% (2015) to 29.59% (2018). Variant classifications of autosomal recessive (AR) variants that previously belonged to VUS or likely pathogenic in the 2015 guidelines were changed toward pathogenic in the 2018 guidelines more frequently than those of autosomal dominant variants (29.17% vs. 6.38%, P = 0.005). Stratification of the PM3 and PP1 rules in the 2018 guidelines led to more substantial escalation than that in the 2015 guidelines. We compared the disease-specific guidelines (2018) with the universal guidelines (2015) using real-world data. Owing to the sophistication of case-level data, the HL-specific guidelines have more explicitly classified AR variants toward "likely pathogenic" or "pathogenic", serving as potential references for other recessive genetic diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Disease-specific ACMG/AMP guidelines improve sequence variant interpretation for hearing loss.Genet Med. 2021 Nov;23(11):2208-2212. doi: 10.1038/s41436-021-01254-2. Epub 2021 Jul 6. Genet Med. 2021. PMID: 34230634 Free PMC article.
-
VIP-HL: Semi-automated ACMG/AMP variant interpretation platform for genetic hearing loss.Hum Mutat. 2021 Dec;42(12):1567-1575. doi: 10.1002/humu.24277. Epub 2021 Sep 2. Hum Mutat. 2021. PMID: 34428318
-
[The updates of the ACMG variant interpretation guidelines affect the pathogenicity determination of OTOF gene variations in patients with auditory neuropathy].Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024 May 7;59(5):455-463. doi: 10.3760/cma.j.cn115330-20231129-00254. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024. PMID: 38811176 Chinese.
-
Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss.Hum Mutat. 2018 Nov;39(11):1593-1613. doi: 10.1002/humu.23630. Hum Mutat. 2018. PMID: 30311386 Free PMC article.
-
Genetic etiology of hereditary hearing loss in the Gulf Cooperation Council countries.Hum Genet. 2022 Apr;141(3-4):595-605. doi: 10.1007/s00439-021-02323-x. Epub 2021 Aug 2. Hum Genet. 2022. PMID: 34338889 Review.
Cited by
-
An overload of missense variants in the OTOG gene may drive a higher prevalence of familial Meniere disease in the European population.Hum Genet. 2024 Mar;143(3):423-435. doi: 10.1007/s00439-024-02643-8. Epub 2024 Mar 22. Hum Genet. 2024. PMID: 38519595 Free PMC article.
-
Explicable prioritization of genetic variants by integration of rule-based and machine learning algorithms for diagnosis of rare Mendelian disorders.Hum Genomics. 2024 Mar 21;18(1):28. doi: 10.1186/s40246-024-00595-8. Hum Genomics. 2024. PMID: 38509596 Free PMC article.
-
GDC: Integration of Multi-Omic and Phenotypic Resources to Unravel the Genetic Pathogenesis of Hearing Loss.Adv Sci (Weinh). 2025 Aug;12(29):e2408891. doi: 10.1002/advs.202408891. Epub 2025 Mar 16. Adv Sci (Weinh). 2025. PMID: 40089864 Free PMC article.
-
Improving detection of monogenic diabetes through reanalysis of GCK variants of uncertain significance.Acta Diabetol. 2025 Aug;62(8):1283-1289. doi: 10.1007/s00592-025-02449-8. Epub 2025 Jan 16. Acta Diabetol. 2025. PMID: 39821340 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

