Loss of FOCAD, operating via the SKI messenger RNA surveillance pathway, causes a pediatric syndrome with liver cirrhosis
- PMID: 35864190
- PMCID: PMC7615854
- DOI: 10.1038/s41588-022-01120-0
Loss of FOCAD, operating via the SKI messenger RNA surveillance pathway, causes a pediatric syndrome with liver cirrhosis
Abstract
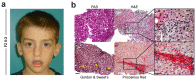
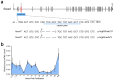
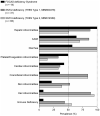
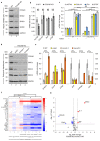
Cirrhosis is usually a late-onset and life-threatening disease characterized by fibrotic scarring and inflammation that disrupts liver architecture and function. While it is typically the result of alcoholism or hepatitis viral infection in adults, its etiology in infants is much less understood. In this study, we report 14 children from ten unrelated families presenting with a syndromic form of pediatric liver cirrhosis. By genome/exome sequencing, we found recessive variants in FOCAD segregating with the disease. Zebrafish lacking focad phenocopied the human disease, revealing a signature of altered messenger RNA (mRNA) degradation processes in the liver. Using patient's primary cells and CRISPR-Cas9-mediated inactivation in human hepatic cell lines, we found that FOCAD deficiency compromises the SKI mRNA surveillance pathway by reducing the levels of the RNA helicase SKIC2 and its cofactor SKIC3. FOCAD knockout hepatocytes exhibited lowered albumin expression and signs of persistent injury accompanied by CCL2 overproduction. Our results reveal the importance of FOCAD in maintaining liver homeostasis and disclose a possible therapeutic intervention point via inhibition of the CCL2/CCR2 signaling axis.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
P.B., A.M.B.-A., V.K., N.O.-H. and S.K. are employed by and receive a salary from Centogene AG (exome sequencing is among the commercially available tests). K.M.G. is founder and director of Suma Genomics (exome sequencing is among the commercially available tests). The remaining authors declare no competing interests.
Figures





Comment in
-
New insights into aetiology of paediatric liver cirrhosis.Nat Rev Gastroenterol Hepatol. 2022 Oct;19(10):623. doi: 10.1038/s41575-022-00676-w. Nat Rev Gastroenterol Hepatol. 2022. PMID: 35974111 No abstract available.
References
-
- Friedman SL. Hepatic fibrosis—overview. Toxicology. 2008;254:120–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

