Comprehensive profiling and characterization of cellular microRNAs in response to coxsackievirus A10 infection in bronchial epithelial cells
- PMID: 35864512
- PMCID: PMC9302563
- DOI: 10.1186/s12985-022-01852-9
Comprehensive profiling and characterization of cellular microRNAs in response to coxsackievirus A10 infection in bronchial epithelial cells
Abstract
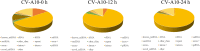
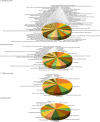
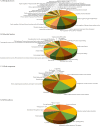
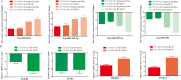
Coxsackievirus A10 (CV-A10), the causative agent of hand, foot, and mouth disease (HFMD), caused a series of outbreaks in recent years and often leads to neurological impairment, but a clear understanding of the disease pathogenesis and host response remains elusive. Cellular microRNAs (miRNAs), a large family of non-coding RNA molecules, have been reported to be key regulators in viral pathogenesis and virus-host interactions. However, the role of host cellular miRNAs defensing against CV-A10 infection is still obscure. To address this issue, we systematically analyzed miRNA expression profiles in CV-A10-infected 16HBE cells by high-throughput sequencing methods in this study. It allowed us to successfully identify 312 and 278 miRNAs with differential expression at 12 h and 24 h post-CV-A10 infection, respectively. Among these, 4 miRNAs and their target genes were analyzed by RT-qPCR, which confirmed the sequencing data. Gene target prediction and enrichment analysis revealed that the predicted targets of these miRNAs were significantly enriched in numerous cellular processes, especially in regulation of basic physical process, host immune response and neurological impairment. And the integrated network was built to further indicate the regulatory roles of miRNAs in host-CV-A10 interactions. Consequently, our findings could provide a beneficial basis for further studies on the regulatory roles of miRNAs relevant to the host immune responses and neuropathogenesis caused by CV-A10 infection.
Keywords: Bioinformatics analysis; Coxsackievirus A10 (CV-A10); Hand, foot, and mouth disease (HFMD); High-throughput sequencing; MicroRNAs (miRNAs).
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

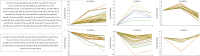

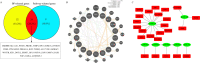
References
-
- Saguil A, Kane SF, Lauters R, Mercado MG. Hand-foot-and-mouth disease: rapid evidence review. Am Fam Physician. 2019;100:408–414. - PubMed
-
- Chang YK, Chen KH, Chen KT. Hand, foot and mouth disease and herpangina caused by enterovirus A71 infections: a review of enterovirus A71 molecular epidemiology, pathogenesis, and current vaccine development. Rev Inst Med Trop Sao Paulo. 2018;60:e70. doi: 10.1590/s1678-9946201860070. - DOI - PMC - PubMed
-
- Kimmis BD, Downing C, Tyring S. Hand-foot-and-mouth disease caused by coxsackievirus A6 on the rise. Cutis. 2018;102:353–356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

