Abnormal antibodies to self-carbohydrates in SARS-CoV-2-infected patients
- PMID: 35865361
- PMCID: PMC9291223
- DOI: 10.1093/pnasnexus/pgac062
Abnormal antibodies to self-carbohydrates in SARS-CoV-2-infected patients
Abstract
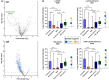
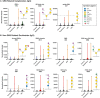
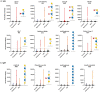
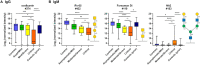
Our immune system is critical for preventing and treating SARS-CoV-2 infections, but aberrant immune responses can have deleterious effects. While antibodies to glycans could recognize the virus and influence the clinical outcome, little is known about their roles. Using a carbohydrate antigen microarray, we profiled serum antibodies in healthy control subjects and COVID-19 patients from two separate cohorts. COVID-19 patients had numerous autoantibodies to self-glycans, including antiganglioside antibodies that can cause neurological disorders. Additionally, nearly all antiglycan IgM signals were lower in COVID-19 patients, indicating a global dysregulation of this class of antibodies. Autoantibodies to certain N-linked glycans correlated with more severe disease, as did low levels of antibodies to the Forssman antigen and ovalbumin. Collectively, this study indicates that expanded testing for antiglycan antibodies could be beneficial for clinical analysis of COVID-19 patients and illustrates the importance of including host and viral carbohydrate antigens when studying immune responses to viruses.
Published by Oxford University Press on behalf of the National Academy of Sciences 2021.
Figures



Update of
-
Abnormal antibodies to self-carbohydrates in SARS-CoV-2 infected patients.bioRxiv [Preprint]. 2020 Oct 16:2020.10.15.341479. doi: 10.1101/2020.10.15.341479. bioRxiv. 2020. Update in: PNAS Nexus. 2022 Jun 17;1(3):pgac062. doi: 10.1093/pnasnexus/pgac062. PMID: 33083799 Free PMC article. Updated. Preprint.
References
-
- Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. 2020. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 324(8):782–793. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

