Evaluating the Microsatellite Instability of Colorectal Cancer Based on Multimodal Deep Learning Integrating Histopathological and Molecular Data
- PMID: 35865460
- PMCID: PMC9295995
- DOI: 10.3389/fonc.2022.925079
Evaluating the Microsatellite Instability of Colorectal Cancer Based on Multimodal Deep Learning Integrating Histopathological and Molecular Data
Abstract
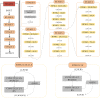
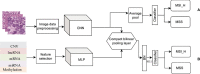
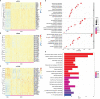
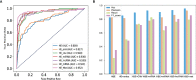
Microsatellite instability (MSI), an important biomarker for immunotherapy and the diagnosis of Lynch syndrome, refers to the change of microsatellite (MS) sequence length caused by insertion or deletion during DNA replication. However, traditional wet-lab experiment-based MSI detection is time-consuming and relies on experimental conditions. In addition, a comprehensive study on the associations between MSI status and various molecules like mRNA and miRNA has not been performed. In this study, we first studied the association between MSI status and several molecules including mRNA, miRNA, lncRNA, DNA methylation, and copy number variation (CNV) using colorectal cancer data from The Cancer Genome Atlas (TCGA). Then, we developed a novel deep learning framework to predict MSI status based solely on hematoxylin and eosin (H&E) staining images, and combined the H&E image with the above-mentioned molecules by multimodal compact bilinear pooling. Our results showed that there were significant differences in mRNA, miRNA, and lncRNA between the high microsatellite instability (MSI-H) patient group and the low microsatellite instability or microsatellite stability (MSI-L/MSS) patient group. By using the H&E image alone, one can predict MSI status with an acceptable prediction area under the curve (AUC) of 0.809 in 5-fold cross-validation. The fusion models integrating H&E image with a single type of molecule have higher prediction accuracies than that using H&E image alone, with the highest AUC of 0.952 achieved when combining H&E image with DNA methylation data. However, prediction accuracy will decrease when combining H&E image with all types of molecular data. In conclusion, combining H&E image with deep learning can predict the MSI status of colorectal cancer, the accuracy of which can further be improved by integrating appropriate molecular data. This study may have clinical significance in practice.
Keywords: H&E images; compact bilinear pooling; microsatellite instability; multi-omics data; multimodal deep learning.
Copyright © 2022 Qiu, Yang, Wang, Yang, Tian, Wang and Yang.
Conflict of interest statement
Authors WQ, JLY, GT, and MY were employed by Geneis Beijing Co., Ltd., Beijing. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Chen W, Zheng R, Zheng S, Ceng H, Zuo T, Jia M. Analysis of Malignant Tumor Incidence and Death in China in 2012. China Cancer (2016) 1):8. doi: 10.11735/j.issn.1004-0242.2015.01.A001 - DOI
-
- Romanowicz-Makowska H, Smolarz B, Langner E, Kozłowska E, Kulig A, Dziki A. Analysis of Microsatellite Instability and BRCA1 Mutations in Patients From Hereditary Nonpolyposis Colorectal Cancer (HNPCC) Family. Pol J Pathol (2005) 56(1):21–6. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

