Initial source of heterogeneity in a model for cell fate decision in the early mammalian embryo
- PMID: 35865503
- PMCID: PMC9184963
- DOI: 10.1098/rsfs.2022.0010
Initial source of heterogeneity in a model for cell fate decision in the early mammalian embryo
Abstract
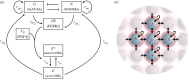
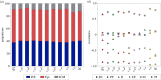
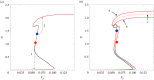
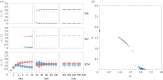
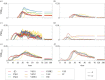
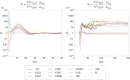
During development, cells from a population of common progenitors evolve towards different fates characterized by distinct levels of specific transcription factors, a process known as cell differentiation. This evolution is governed by gene regulatory networks modulated by intercellular signalling. In order to evolve towards distinct fates, cells forming the population of common progenitors must display some heterogeneity. We applied a modelling approach to obtain insights into the possible sources of cell-to-cell variability initiating the specification of cells of the inner cell mass into epiblast or primitive endoderm cells in early mammalian embryo. At the single-cell level, these cell fates correspond to three possible steady states of the model. A combination of numerical simulations and bifurcation analyses predicts that the behaviour of the model is preserved with respect to the source of variability and that cell-cell coupling induces the emergence of multiple steady states associated with various cell fate configurations, and to a distribution of the levels of expression of key transcription factors. Statistical analysis of these time-dependent distributions reveals differences in the evolutions of the variance-to-mean ratios of key variables of the system, depending on the simulated source of variability, and, by comparison with experimental data, points to the rate of synthesis of the key transcription factor NANOG as a likely initial source of heterogeneity.
Keywords: bifurcation; cell differentiation; noise; probability distribution; tristability; variability.
© 2022 The Author(s).
Figures


References
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
