RGD-Labeled Hemocytes With High Migration Activity Display a Potential Immunomodulatory Role in the Pacific Oyster Crassostrea gigas
- PMID: 35865522
- PMCID: PMC9294365
- DOI: 10.3389/fimmu.2022.914899
RGD-Labeled Hemocytes With High Migration Activity Display a Potential Immunomodulatory Role in the Pacific Oyster Crassostrea gigas
Abstract
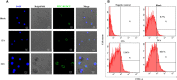
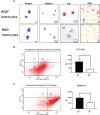
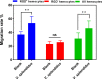
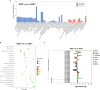
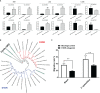
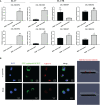
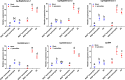
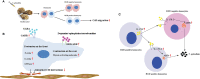
Immunocyte migration to infection sites is important for host cellular defense, but the main types of migrating hemocytes and their mechanisms against pathogen invasions are unclear in invertebrates. In the present study, a population of hemocytes in the Pacific oyster Crassostrea gigas labeled with a fluorescein isothiocyanate (FITC)-conjugated Arg-Gly-Asp (RGD)-containing peptide was sorted. RGD+ hemocytes were characterized by a smaller cell size and cytoplasmic-nucleo ratio, fewer cytoplasmic granules, and higher levels of myeloperoxidase, reactive oxygen species, and intracellular free calcium concentration. RGD+ hemocytes exhibited a high level of migration activity, which was further induced after V. splendidus infection. Transcriptome analysis revealed that RGD+ hemocytes highly expressed a series of migration-related genes, which together with migration-promoting genes were significantly upregulated after V. splendidus infection. The neuroendocrine system was also proven to regulate the migration activity of RGD+ hemocytes, especially with the excitatory neuroendocrine factor dopamine, which promoted migration activity as confirmed by receptor blocking assays. Meanwhile, RGD+ hemocytes could highly express immunomodulatory factor interleukin (IL)-17s and their receptor genes, which was positively related to the production of antimicrobial peptides in whole hemocytes after V. splendidus infection. Collectively, this study identified a specific hemocyte population, i.e., RGD+ hemocytes, that shows high migration activity in response to pathogen infection and exerts a potential immunomodulatory role by highly expressing IL-17s that might enhance the hemocytes' antimicrobial peptide production in oysters.
Keywords: C. gigas; RGD labeled hemocytes; antimicrobial immunity; immunomodulatory; migration activity.
Copyright © 2022 Lv, Qiu, Wang, Liu, Liu, Wang and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The activated β-integrin (CgβV) enhances RGD-binding and phagocytic capabilities of hemocytes in Crassostrea gigas.Fish Shellfish Immunol. 2019 Apr;87:638-649. doi: 10.1016/j.fsi.2019.01.047. Epub 2019 Feb 10. Fish Shellfish Immunol. 2019. PMID: 30753917
-
The specifically enhanced cellular immune responses in Pacific oyster (Crassostrea gigas) against secondary challenge with Vibrio splendidus.Dev Comp Immunol. 2014 Jul;45(1):141-50. doi: 10.1016/j.dci.2014.02.015. Epub 2014 Mar 6. Dev Comp Immunol. 2014. PMID: 24607288
-
The hematopoiesis in gill and its role in the immune response of Pacific oyster Crassostrea gigas against secondary challenge with Vibrio splendidus.Dev Comp Immunol. 2017 Jun;71:59-69. doi: 10.1016/j.dci.2017.01.024. Epub 2017 Feb 1. Dev Comp Immunol. 2017. PMID: 28159592
-
The sensing pattern and antitoxic response of Crassostrea gigas against extracellular products of Vibrio splendidus.Dev Comp Immunol. 2020 Jan;102:103467. doi: 10.1016/j.dci.2019.103467. Epub 2019 Aug 16. Dev Comp Immunol. 2020. PMID: 31425720
-
The new insights into the oyster antimicrobial defense: Cellular, molecular and genetic view.Fish Shellfish Immunol. 2015 Sep;46(1):50-64. doi: 10.1016/j.fsi.2015.02.040. Epub 2015 Mar 7. Fish Shellfish Immunol. 2015. PMID: 25753917 Review.
Cited by
-
Hallmarks of crustacean immune hemocytes at single-cell resolution.Front Immunol. 2023 Jan 24;14:1121528. doi: 10.3389/fimmu.2023.1121528. eCollection 2023. Front Immunol. 2023. PMID: 36761772 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

