Chromatin Ubiquitination Guides DNA Double Strand Break Signaling and Repair
- PMID: 35865631
- PMCID: PMC9294282
- DOI: 10.3389/fcell.2022.928113
Chromatin Ubiquitination Guides DNA Double Strand Break Signaling and Repair
Abstract
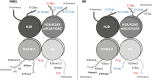
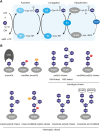
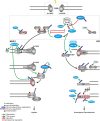
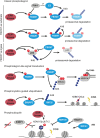
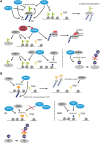
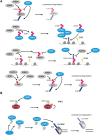
Chromatin is the context for all DNA-based molecular processes taking place in the cell nucleus. The initial chromatin structure at the site of the DNA damage determines both, lesion generation and subsequent activation of the DNA damage response (DDR) pathway. In turn, proceeding DDR changes the chromatin at the damaged site and across large fractions of the genome. Ubiquitination, besides phosphorylation and methylation, was characterized as an important chromatin post-translational modification (PTM) occurring at the DNA damage site and persisting during the duration of the DDR. Ubiquitination appears to function as a highly versatile "signal-response" network involving several types of players performing various functions. Here we discuss how ubiquitin modifiers fine-tune the DNA damage recognition and response and how the interaction with other chromatin modifications ensures cell survival.
Keywords: DNA damage response; DNA repair; chromatin; double-strand breaks; ubiquitination.
Copyright © 2022 Kolobynina, Rapp and Cardoso.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

