Emerging Roles of Long Noncoding RNAs in Breast Cancer Epigenetics and Epitranscriptomics
- PMID: 35865634
- PMCID: PMC9294602
- DOI: 10.3389/fcell.2022.922351
Emerging Roles of Long Noncoding RNAs in Breast Cancer Epigenetics and Epitranscriptomics
Abstract
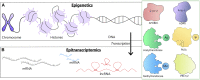
Breast carcinogenesis is a multistep process that involves both genetic and epigenetic changes. Epigenetics refers to reversible changes in gene expression that are not accompanied by changes in gene sequence. In breast cancer (BC), dysregulated epigenetic changes, such as DNA methylation and histone modifications, are accompanied by epitranscriptomic changes, in particular adenine to inosine modifications within RNA molecules. Factors that trigger these phenomena are largely unknown, but there is evidence for widespread participation of long noncoding RNAs (lncRNAs) that already have been linked to virtually any aspect of BC biology, making them promising biomarkers and therapeutic targets in BC patients. Here, we provide a systematic review of known and possible roles of lncRNAs in epigenetic and epitranscriptomic processes, along with methods and tools to study them, followed by a brief overview of current challenges regarding the use of lncRNAs in medical applications.
Keywords: RNA modifications; breast cancer; epigenetics; epitranscriptomics; long noncoding RNAs.
Copyright © 2022 Wanowska, Samorowska and Szcześniak.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Andresini O., Rossi M. N., Matteini F., Petrai S., Santini T., Maione R. (2019). The Long Non-Coding RNA Kcnq1ot1 Controls Maternal P57 Expression in Muscle Cells by Promoting H3K27me3 Accumulation to an Intragenic MyoD-Binding Region. Epigenetics Chromatin 12 (1), 8. 10.1186/s13072-019-0253-1 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

