Isolation and Characterization of vB_kpnM_17-11, a Novel Phage Efficient Against Carbapenem-Resistant Klebsiella pneumoniae
- PMID: 35865823
- PMCID: PMC9294173
- DOI: 10.3389/fcimb.2022.897531
Isolation and Characterization of vB_kpnM_17-11, a Novel Phage Efficient Against Carbapenem-Resistant Klebsiella pneumoniae
Abstract
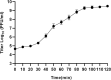
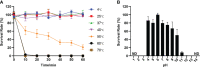
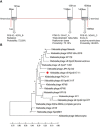
Phages and phage-encoded proteins exhibit promising prospects in the treatment of Carbapenem-Resistant Klebsiella pneumoniae (CRKP) infections. In this study, a novel Klebsiella pneumoniae phage vB_kpnM_17-11 was isolated and identified by using a CRKP host. vB_kpnM_17-11 has an icosahedral head and a retractable tail. The latent and exponential phases were 30 and 60 minutes, respectively; the burst size was 31.7 PFU/cell and the optimal MOI was 0.001. vB_kpnM_17-11 remained stable in a wide range of pH (4-8) and temperature (4-40°C). The genome of vB_kpnM_17-11 is 165,894 bp, double-stranded DNA (dsDNA), containing 275 Open Reading Frames (ORFs). It belongs to the family of Myoviridae, order Caudovirales, and has a close evolutionary relationship with Klebsiella phage PKO111. Sequence analysis showed that the 4530 bp orf022 of vB_kpnM_17-11 encodes a putative depolymerase. In vitro testing demonstrated that vB_kpnM_17-11 can decrease the number of K. pneumoniae by 105-fold. In a mouse model of infection, phage administration improved survival and reduced the number of K. pneumoniae in the abdominal cavity by 104-fold. In conclusion, vB_kpnM_17-11 showed excellent in vitro and in vivo performance against K. pneumoniae infection and constitutes a promising candidate for the development of phage therapy against CRKP.
Keywords: Klebsiella pneumonia; animal model; depolymerase; phage; phage therapy.
Copyright © 2022 Bai, Zhang, Liang, Chen, Wang, Wang, Martín-Rodríguez, Sjöling, Hu and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Isolation, Identification, and Biological Characterization of Phage vB_KpnM_KpVB3 Targeting Carbapenem-Resistant Klebsiella pneumoniae ST11.J Glob Antimicrob Resist. 2024 Jun;37:179-184. doi: 10.1016/j.jgar.2024.03.007. Epub 2024 Mar 30. J Glob Antimicrob Resist. 2024. PMID: 38561142
-
Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB_KshKPC-M against carbapenemase-producing Klebsiella pneumoniae strains (CRKP) isolated from Ventilator-associated pneumoniae (VAP) infection of COVID-19 patients.Ann Clin Microbiol Antimicrob. 2023 Feb 24;22(1):18. doi: 10.1186/s12941-023-00567-1. Ann Clin Microbiol Antimicrob. 2023. PMID: 36829156 Free PMC article.
-
Identification of a novel phage depolymerase against ST11 K64 carbapenem-resistant Klebsiella pneumoniae and its therapeutic potential.J Bacteriol. 2025 Apr 17;207(4):e0038724. doi: 10.1128/jb.00387-24. Epub 2025 Mar 26. J Bacteriol. 2025. PMID: 40135928 Free PMC article.
-
Phenotypic and Genomic Comparison of Klebsiella pneumoniae Lytic Phages: vB_KpnM-VAC66 and vB_KpnM-VAC13.Viruses. 2021 Dec 21;14(1):6. doi: 10.3390/v14010006. Viruses. 2021. PMID: 35062209 Free PMC article.
-
Characterization of a new lytic bacteriophage (vB_KpnM_KP1) targeting Klebsiella pneumoniae strains associated with nosocomial infections.Virology. 2025 Jun;607:110526. doi: 10.1016/j.virol.2025.110526. Epub 2025 Apr 4. Virology. 2025. PMID: 40203466
Cited by
-
Potential of an Isolated Bacteriophage to Inactivate Klebsiella pneumoniae: Preliminary Studies to Control Urinary Tract Infections.Antibiotics (Basel). 2024 Feb 19;13(2):195. doi: 10.3390/antibiotics13020195. Antibiotics (Basel). 2024. PMID: 38391581 Free PMC article.
-
Bacteriophage-derived depolymerase: a review on prospective antibacterial agents to combat Klebsiella pneumoniae.Arch Virol. 2025 Mar 8;170(4):70. doi: 10.1007/s00705-025-06257-x. Arch Virol. 2025. PMID: 40057622 Review.
-
Analysis of a novel phage as a promising biological agent targeting multidrug resistant Klebsiella pneumoniae.AMB Express. 2025 Mar 5;15(1):37. doi: 10.1186/s13568-025-01846-0. AMB Express. 2025. PMID: 40044971 Free PMC article.
-
In Vitro Activity, Stability and Molecular Characterization of Eight Potent Bacteriophages Infecting Carbapenem-Resistant Klebsiella pneumoniae.Viruses. 2022 Dec 30;15(1):117. doi: 10.3390/v15010117. Viruses. 2022. PMID: 36680156 Free PMC article.
-
Characterization of a Lytic Bacteriophage and Demonstration of Its Combined Lytic Effect with a K2 Depolymerase on the Hypervirulent Klebsiella pneumoniae Strain 52145.Microorganisms. 2023 Mar 6;11(3):669. doi: 10.3390/microorganisms11030669. Microorganisms. 2023. PMID: 36985241 Free PMC article.
References
-
- Amarillas L., Rubí-Rangel L., Chaidez C., González-Robles A., Lightbourn-Rojas L., León-Félix J. (2017). Isolation and Characterization of phiLLS, a Novel Phage With Potential Biocontrol Agent Against Multidrug-Resistant Escherichia Coli. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01355 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

