Combating small molecule environmental contaminants: detection and sequestration using functional nucleic acids
- PMID: 35865900
- PMCID: PMC9258336
- DOI: 10.1039/d2sc00117a
Combating small molecule environmental contaminants: detection and sequestration using functional nucleic acids
Abstract
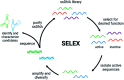
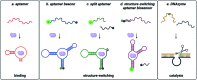
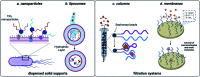
Small molecule contaminants pose a significant threat to the environment and human health. While regulations are in place for allowed limits in many countries, detection and remediation of contaminants in more resource-limited settings and everyday environmental sources remains a challenge. Functional nucleic acids, including aptamers and DNA enzymes, have emerged as powerful options for addressing this challenge due to their ability to non-covalently interact with small molecule targets. The goal of this perspective is to outline recent efforts toward the selection of aptamers for small molecules and describe their subsequent implementation for environmental applications. Finally, we provide an outlook that addresses barriers that hinder these technologies from being widely adopted in field friendly settings and propose a path forward toward addressing these challenges.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


References
-
- Pool R. and Rusch E., Identifying and Reducing Environmental Health Risks of Chemicals in Our Society, 2014 - PubMed
-
- D'Surney S. J. and Smith M. D., Encycl. Toxicol., 2005, pp. 526–530
-
- Xu Y. Hassan M. M. Sharma A. S. Li H. Chen Q. Crit. Rev. Food Sci. Nutr. 2021:1–19.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

