Integrated analysis of the clinical consequence and associated gene expression of ALK in ALK-positive human cancers
- PMID: 35865984
- PMCID: PMC9293659
- DOI: 10.1016/j.heliyon.2022.e09878
Integrated analysis of the clinical consequence and associated gene expression of ALK in ALK-positive human cancers
Abstract
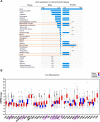
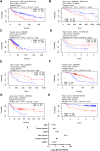
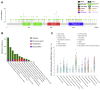
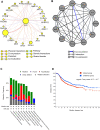
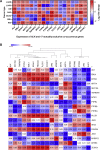
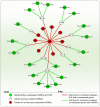
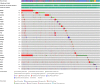
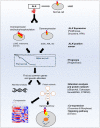
Anaplastic lymphoma kinase (ALK) is a tyrosine kinase receptor that is genetically altered in several cancers, including NSCLC, melanoma, lymphoma, and other tumors. Although ALK is associated with various cancers, the relationship between ALK expression and patient prognosis in different cancers is poorly understood. Here, using multidimensional approaches, we revealed the correlation between ALK expression and the clinical outcomes of patients with LUAD, melanoma, OV, DLBC, AML, and BC. We analyzed ALK transcriptional expression, patient survival rate, genetic alteration, protein network, and gene and microRNA (miRNA) co-expression. Compared to that in normal tissues, higher ALK expression was found in LUAD, melanoma, and OV, which are associated with poor patient survival rates. In contrast, lower transcriptional expression was found to decrease the survival rate of patients with DLBC, AML, and BC. A total of 202 missense mutations, 17 truncating mutations, 7 fusions, and 3 in-frame mutations were identified. Further, 17 genes and 19 miRNAs were found to be exclusively co-expressed and echinoderm microtubule-associated protein-like 4 (EML4) was identified as the most positively correlated gene (log odds ratio >3). The gene ontology and signaling pathways of the genes co-expressed with ALK in these six cancers were also identified. Our findings offer a basis for ALK as a prognostic biomarker and therapeutic target in cancers, which will potentially contribute to precision oncology and assist clinicians in identifying suitable treatment options.
Keywords: ALK expression; Cancers; Co-expression; LUAD; Patient prognosis.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
miR-100-5p confers resistance to ALK tyrosine kinase inhibitors Crizotinib and Lorlatinib in EML4-ALK positive NSCLC.Biochem Biophys Res Commun. 2019 Apr 2;511(2):260-265. doi: 10.1016/j.bbrc.2019.02.016. Epub 2019 Feb 18. Biochem Biophys Res Commun. 2019. PMID: 30791979
-
Going beneath the tip of the iceberg. Identifying and understanding EML4-ALK variants and TP53 mutations to optimize treatment of ALK fusion positive (ALK+) NSCLC.Lung Cancer. 2021 Aug;158:126-136. doi: 10.1016/j.lungcan.2021.06.012. Epub 2021 Jun 12. Lung Cancer. 2021. PMID: 34175504 Review.
-
Concomitant mutation status of ALK-rearranged non-small cell lung cancers and its prognostic impact on patients treated with crizotinib.Transl Lung Cancer Res. 2021 Mar;10(3):1525-1535. doi: 10.21037/tlcr-21-160. Transl Lung Cancer Res. 2021. PMID: 33889527 Free PMC article.
-
Distribution of EML4-ALK fusion variants and clinical outcomes in patients with resected non-small cell lung cancer.Lung Cancer. 2020 Nov;149:154-161. doi: 10.1016/j.lungcan.2020.09.012. Epub 2020 Sep 24. Lung Cancer. 2020. PMID: 33017727
-
ALK inhibitors in the treatment of advanced NSCLC.Cancer Treat Rev. 2014 Mar;40(2):300-6. doi: 10.1016/j.ctrv.2013.07.002. Epub 2013 Aug 7. Cancer Treat Rev. 2014. PMID: 23931927 Review.
Cited by
-
Neuronal ALKAL2 and its ALK receptor contribute to the development of colitis-associated colorectal cancer.Proc Natl Acad Sci U S A. 2025 Jun 17;122(24):e2500632122. doi: 10.1073/pnas.2500632122. Epub 2025 Jun 10. Proc Natl Acad Sci U S A. 2025. PMID: 40493183
-
A humanized anaplastic lymphoma kinase (ALK)-directed antibody-drug conjugate with pyrrolobenzodiazepine payload demonstrates efficacy in ALK-expressing cancers.Nat Commun. 2025 Aug 15;16(1):7578. doi: 10.1038/s41467-025-62979-1. Nat Commun. 2025. PMID: 40813394 Free PMC article.
References
-
- Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S.-i., Watanabe H., Kurashina K., Hatanaka H., et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. - PubMed
-
- Saifullah, Tsukahara T. Integrated analysis of ALK higher expression in human cancer and downregulation in LUAD using RNA molecular scissors. Clin. Transl. Oncol. 2022 - PubMed
-
- Morris S., Kirstein M., Valentine M., Dittmer K., Shapiro D., Saltman D., Look A. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. - PubMed
LinkOut - more resources
Full Text Sources

