Interpretation of anomalously long crosslinks in ribosome crosslinking reveals the ribosome interaction in stationary phase E. coli
- PMID: 35866168
- PMCID: PMC9257603
- DOI: 10.1039/d2cb00101b
Interpretation of anomalously long crosslinks in ribosome crosslinking reveals the ribosome interaction in stationary phase E. coli
Abstract
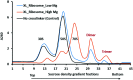
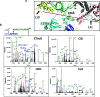
Crosslinking mass spectrometry (XL-MS) of bacterial ribosomes revealed the dynamic intra- and intermolecular interactions within the ribosome structure. It has been also extended to capture the interactions of ribosome binding proteins during translation. Generally, XL-MS often identified the crosslinks within a cross-linkable distance (<40 Å) using amine-reactive crosslinkers. The crosslinks larger than cross-linkable distance (>40 Å) are always difficult to interpret and remain unnoticed. Here, we focused on stationary phase bacterial ribosome crosslinking that yields ultra-long crosslinks in an E. coli cell lysate. We explain these ultra-long crosslinks with the combination of sucrose density gradient centrifugation, chemical crosslinking, high-resolution mass spectrometry, and electron microscopy analysis. Multiple ultra-long crosslinks were observed in E. coli ribosomes for example ribosomal protein L19 (K63, K94) crosslinks with L21 (K71, K81) at two locations that are about 100 Å apart. Structural mapping of such ultra-long crosslinks in 70S ribosomes suggested that these crosslinks are not potentially formed within one 70S particle and could be a result of dimer and trimer formation as evidenced by negative staining electron microscopy. Ribosome dimerization captured by chemical crosslinking reaction could be an indication of ribosome-ribosome interactions in the stationary phase.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
Determination of HER2 binding domain in antigen-antibody complexes based on chemical crosslinking mass spectrometry.J Proteomics. 2023 Aug 30;286:104954. doi: 10.1016/j.jprot.2023.104954. Epub 2023 Jun 28. J Proteomics. 2023. PMID: 37390893
-
A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.Nat Commun. 2017 Sep 28;8(1):722. doi: 10.1038/s41467-017-00718-x. Nat Commun. 2017. PMID: 28959045 Free PMC article.
-
Structural analysis of 70S ribosomes by cross-linking/mass spectrometry reveals conformational plasticity.Sci Rep. 2020 Jul 28;10(1):12618. doi: 10.1038/s41598-020-69313-3. Sci Rep. 2020. PMID: 32724211 Free PMC article.
-
Protein structure dynamics by crosslinking mass spectrometry.Curr Opin Struct Biol. 2023 Jun;80:102599. doi: 10.1016/j.sbi.2023.102599. Epub 2023 Apr 25. Curr Opin Struct Biol. 2023. PMID: 37104977 Review.
-
Insights into protein biosynthesis from structures of bacterial ribosomes.Curr Opin Struct Biol. 2007 Jun;17(3):302-9. doi: 10.1016/j.sbi.2007.05.009. Epub 2007 Jun 15. Curr Opin Struct Biol. 2007. PMID: 17574829 Review.
Cited by
-
Cross-linking mass spectrometry for mapping protein complex topologies in situ.Essays Biochem. 2023 Mar 29;67(2):215-228. doi: 10.1042/EBC20220168. Essays Biochem. 2023. PMID: 36734207 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

