Rational design of a dual-reactive probe for imaging the biogenesis of both H2S and GSH from l-Cys rather than d-Cys in live cells
- PMID: 35866170
- PMCID: PMC9257618
- DOI: 10.1039/d2cb00105e
Rational design of a dual-reactive probe for imaging the biogenesis of both H2S and GSH from l-Cys rather than d-Cys in live cells
Abstract
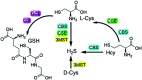
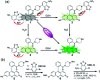
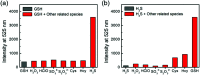
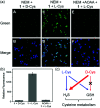
Biothiols and their interconversion are involved in cellular redox homeostasis as well as many physiological processes. Here, a dual-reactive dual-quenching fluorescent probe was rationally developed based on thiolysis reactions of 7-nitrobenzoxadiazole (NBD) tertiary amine and 7-cyanobenzoxadiazole (CBD) arylether for imaging of the biothiol interconversion. We demonstrate that the NBD-CBD probe exhibits very weak background fluorescence due to the dual-quenching effects, and can be dual-activated by H2S and GSH with an over 500-fold fluorescence increase at 525 nm. In addition, the probe shows high sensitivity, excellent selectivity, and good biocompatibility, all of which promote the simultaneous detection of both H2S and GSH in live cells. Importantly, probe 1 was successfully employed to reveal the biogenesis of both H2S and GSH from l-Cys rather than from d-Cys, and therefore, d-Cys would be solely converted into H2S, which may help understand the more H2S generation from d-Cys than from l-Cys in live cells.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
-
- Yang G. Wu L. Jiang B. Yang W. Qi J. Cao K. Meng Q. Mustafa A. K. Mu W. Zhang S. Snyder S. H. Wang R. Science. 2008;332:587. doi: 10.1126/science.1162667. - DOI - PMC - PubMed
- Cao X. Ding L. Xie Z. Yang Y. Whiteman M. Moore P. K. Bian J.-S. Antioxid. Redox Signaling. 2019;31:1–38. doi: 10.1089/ars.2017.7058. - DOI - PMC - PubMed
- Filipovic M. R. Zivanovic J. Alvarez B. Banerjee R. Chem. Rev. 2018;118:1253–1337. doi: 10.1021/acs.chemrev.7b00205. - DOI - PMC - PubMed
- Trujillo M. Alvarez B. Radi R. Free Radical Res. 2016;50:150–171. doi: 10.3109/10715762.2015.1089988. - DOI - PubMed
- Rabe von Pappenheim F. Wensien M. Ye J. Uranga J. Irisarri I. de Vries J. Funk L.-M. Mata R. A. Tittmann K. Nat. Chem. Biol. 2022;18:368–375. doi: 10.1038/s41589-021-00966-5. - DOI - PMC - PubMed
- Bora P. Manna S. Nair M. A. Sathe R. R.-M. Singh S. Sreyas Adury V. S. Gupta K. Mukherjee A. Saini D. K. Kamat S. S. Hazra A. B. Chakrapani H. Chem. Sci. 2021;12:12939–12949. doi: 10.1039/D1SC03828A. - DOI - PMC - PubMed
- Levinn C. M. Cerda M. M. Pluth M. D. Acc. Chem. Res. 2019;52:2723–2731. doi: 10.1021/acs.accounts.9b00315. - DOI - PMC - PubMed
- Ni X. Kelly S. S. Xu S. Xian M. Acc. Chem. Res. 2021;54:3968–3978. doi: 10.1021/acs.accounts.1c00506. - DOI - PubMed
-
- Wang R. Physiol. Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. - DOI - PubMed
- Szabo C. Biochem. Pharmacol. 2018;149:5–19. doi: 10.1016/j.bcp.2017.09.010. - DOI - PMC - PubMed
- Liu Y. Lu M. Hu L. Wong P. T. Webb G. D. Bian J. Antioxid. Redox Signaling. 2012;17:141–185. doi: 10.1089/ars.2011.4005. - DOI - PubMed
- Modis K. Ju Y. Ahmad A. Untereiner A. A. Altaany Z. Wu L. Szabo C. Wang R. Pharmacol. Res. 2016;113:116–124. doi: 10.1016/j.phrs.2016.08.023. - DOI - PMC - PubMed
- Khodade V. S. Aggarwal S. C. Pharoah B. M. Paolocci N. Toscano J. P. Chem. Sci. 2021;12:8252–8259. doi: 10.1039/D1SC01550H. - DOI - PMC - PubMed
-
- Yang K. Cao F. Tao L. Zhu Y. Xue Y. Front. Physiol. 2022;13:840293. doi: 10.3389/fphys.2022.840293. - DOI - PMC - PubMed
- Liu T. Sun L. Zhang Y. Wang Y. Zheng J. J. Biochem. Mol. Toxicol. 2022;36:e22942. - PubMed
- Cairns R. A. Harris I. S. Mak T. W. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. - DOI - PubMed
- Hwang C. Sinskey A. J. Lodish H. F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. - DOI - PubMed
-
- Kimura H. Amino Acids. 2011;41:113. doi: 10.1007/s00726-010-0510-x. - DOI - PubMed
- Shibuya N. Koike S. Tanaka M. Ishigami-Yuasa M. Kimura Y. Ogasawara Y. Fukui K. Nagahara N. Kimura H. Nat. Commun. 2013;4:1366. doi: 10.1038/ncomms2371. - DOI - PubMed
- Paul B. D. Sbodio J. I. Snyder S. H. Trends Pharmacol. Sci. 2018;39:513–524. doi: 10.1016/j.tips.2018.02.007. - DOI - PMC - PubMed
- Lu S. C. Biochim. Biophys. Acta, Gen. Subj. 2013;1830:3143–3153. doi: 10.1016/j.bbagen.2012.09.008. - DOI - PMC - PubMed
- Stipanuk M. H. Annu. Rev. Nutr. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. - DOI - PubMed
-
- Szabo C. Nat. Rev. Drug Discovery. 2016;15:185–203. doi: 10.1038/nrd.2015.1. - DOI - PMC - PubMed
- Szabo C. Papapetropoulos A. Pharmacol. Rev. 2017;69:497–564. doi: 10.1124/pr.117.014050. - DOI - PMC - PubMed
- Dorszewska J. Prendecki M. Oczkowska A. Dezor M. Kozubski W. Curr. Alzheimer Res. 2016;13:952–963. doi: 10.2174/1567205013666160314150501. - DOI - PubMed
- Kennedy L. Sandhu J. K. Harper M. Cuperlovic-Culf M. Biomolecules. 2020;10:1429. doi: 10.3390/biom10101429. - DOI - PMC - PubMed
- Coletta C. Modis K. Szczesny B. Brunyanszki A. Olah G. Rios E. C.-S. Yanagi K. Ahmad A. Papapetropoulos A. Szabo C. Mol. Med. 2015;21:1–14. doi: 10.2119/molmed.2015.00035. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

