Listeria monocytogenes utilizes the ClpP1/2 proteolytic machinery for fine-tuned substrate degradation at elevated temperatures
- PMID: 35866172
- PMCID: PMC9257651
- DOI: 10.1039/d2cb00077f
Listeria monocytogenes utilizes the ClpP1/2 proteolytic machinery for fine-tuned substrate degradation at elevated temperatures
Abstract
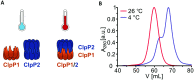
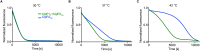
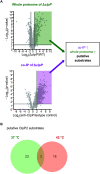
Listeria monocytogenes exhibits two ClpP isoforms (ClpP1/ClpP2) which assemble into a heterooligomeric complex with enhanced proteolytic activity. Herein, we demonstrate that the formation of this complex depends on temperature and reaches a maximum ratio of about 1 : 1 at 30 °C, while almost no complex formation occurred below 4 °C. In order to decipher the role of the two isoforms at elevated temperatures, we constructed L. monocytogenes ClpP1, ClpP2 and ClpP1/2 knockout strains and analyzed their protein regulation in comparison to the wild type (WT) strain via whole proteome mass-spectrometry (MS) at 37 °C and 42 °C. While the ΔclpP1 strain only altered the expression of very few proteins, the ΔclpP2 and ΔclpP1/2 strains revealed the dysregulation of many proteins at both temperatures. These effects were corroborated by crosslinking co-immunoprecipitation MS analysis. Thus, while ClpP1 serves as a mere enhancer of protein degradation in the heterocomplex, ClpP2 is essential for ClpX binding and functions as a gatekeeper for substrate entry. Applying an integrated proteomic approach combining whole proteome and co-immunoprecipitation datasets, several putative ClpP2 substrates were identified in the context of different temperatures and discussed with regards to their function in cellular pathways such as the SOS response.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures






Similar articles
-
Assembly and proteolytic processing of mycobacterial ClpP1 and ClpP2.BMC Biochem. 2011 Dec 1;12:61. doi: 10.1186/1471-2091-12-61. BMC Biochem. 2011. PMID: 22132756 Free PMC article.
-
Distinct clpP genes control specific adaptive responses in Bacillus thuringiensis.J Bacteriol. 2002 Oct;184(20):5554-62. doi: 10.1128/JB.184.20.5554-5562.2002. J Bacteriol. 2002. PMID: 12270812 Free PMC article.
-
Two Isoforms of Clp Peptidase in Pseudomonas aeruginosa Control Distinct Aspects of Cellular Physiology.J Bacteriol. 2017 Jan 12;199(3):e00568-16. doi: 10.1128/JB.00568-16. Print 2017 Feb 1. J Bacteriol. 2017. PMID: 27849175 Free PMC article.
-
Identification of the inhibitory mechanism of ecumicin and rufomycin 4-7 on the proteolytic activity of Mycobacterium tuberculosis ClpC1/ClpP1/ClpP2 complex.Tuberculosis (Edinb). 2023 Jan;138:102298. doi: 10.1016/j.tube.2022.102298. Epub 2022 Dec 24. Tuberculosis (Edinb). 2023. PMID: 36580851 Free PMC article.
-
Chemical Cross-Linking Enables Drafting ClpXP Proximity Maps and Taking Snapshots of In Situ Interaction Networks.Cell Chem Biol. 2019 Jan 17;26(1):48-59.e7. doi: 10.1016/j.chembiol.2018.10.007. Epub 2018 Nov 8. Cell Chem Biol. 2019. PMID: 30415967
Cited by
-
ClpP2 proteasomes and SpxA1 determine Listeria monocytogenes tartrolon B hyper-resistance.PLoS Genet. 2025 Apr 4;21(4):e1011621. doi: 10.1371/journal.pgen.1011621. eCollection 2025 Apr. PLoS Genet. 2025. PMID: 40184427 Free PMC article.
-
The essential M. tuberculosis Clp protease is functionally asymmetric in vivo.Sci Adv. 2022 May 6;8(18):eabn7943. doi: 10.1126/sciadv.abn7943. Epub 2022 May 4. Sci Adv. 2022. PMID: 35507665 Free PMC article.
-
High density genomic surveillance and risk profiling of clinical Listeria monocytogenes subtypes in Germany.Genome Med. 2024 Oct 7;16(1):115. doi: 10.1186/s13073-024-01389-2. Genome Med. 2024. PMID: 39375806 Free PMC article.
-
Structure-guided development of selective caseinolytic protease P agonists as antistaphylococcal agents.Cell Rep Med. 2024 Dec 17;5(12):101837. doi: 10.1016/j.xcrm.2024.101837. Epub 2024 Nov 29. Cell Rep Med. 2024. PMID: 39615486 Free PMC article.
-
Stress Adaptation Responses of a Listeria monocytogenes 1/2a Strain via Proteome Profiling.Foods. 2023 May 27;12(11):2166. doi: 10.3390/foods12112166. Foods. 2023. PMID: 37297410 Free PMC article.
References
LinkOut - more resources
Full Text Sources

