Differentially Active and Conserved Neural Enhancers Define Two Forms of Adaptive Noncoding Evolution in Humans
- PMID: 35866592
- PMCID: PMC9348619
- DOI: 10.1093/gbe/evac108
Differentially Active and Conserved Neural Enhancers Define Two Forms of Adaptive Noncoding Evolution in Humans
Abstract
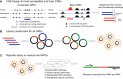
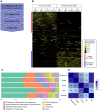
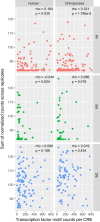
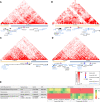
The human and chimpanzee genomes are strikingly similar, but our neural phenotypes are very different. Many of these differences are likely driven by changes in gene expression, and some of those changes may have been adaptive during human evolution. Yet, the relative contributions of positive selection on regulatory regions or other functional regulatory changes are unclear. Where are these changes located throughout the human genome? Are functional regulatory changes near genes or are they in distal enhancer regions? In this study, we experimentally combined both human and chimpanzee cis-regulatory elements (CREs) that showed either (1) signs of accelerated evolution in humans or (2) that have been shown to be active in the human brain. Using a massively parallel reporter assay, we tested the ability of orthologous human and chimpanzee CREs to activate transcription in induced pluripotent stem-cell-derived neural progenitor cells and neurons. With this assay, we identified 179 CREs with differential activity between human and chimpanzee; in contrast, we found 722 CREs with signs of positive selection in humans. Selection and differentially expressed CREs strikingly differ in level of expression, size, and genomic location. We found a subset of 69 CREs in loci with genetic variants associated with neuropsychiatric diseases, which underscores the consequence of regulatory activity in these loci for proper neural development and function. By combining CREs that either experienced recent selection in humans or CREs that are functional brain enhancers, presents a novel way of studying the evolution of noncoding elements that contribute to human neural phenotypes.
Keywords: Cis-regulation; human brain evolution; induced pluripotent stem cells; massively parallel enhancer assay.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Babarinde IA, Saitou N. 2016. Genomic locations of conserved noncoding sequences and their proximal protein-coding genes in mammalian expression dynamics. Mol Biol Evol. 33:1807–1817. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

