KSHV Topologically Associating Domains in Latent and Reactivated Viral Chromatin
- PMID: 35867573
- PMCID: PMC9327698
- DOI: 10.1128/jvi.00565-22
KSHV Topologically Associating Domains in Latent and Reactivated Viral Chromatin
Abstract
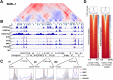
Eukaryotic genomes are structurally organized via the formation of multiple loops that create gene expression regulatory units called topologically associating domains (TADs). Here we revealed the KSHV TAD structure at 500 bp resolution and constructed a 3D KSHV genomic structural model with 2 kb binning. The latent KSHV genome formed very similar genomic architectures in three different naturally infected PEL cell lines and in an experimentally infected epithelial cell line. The majority of the TAD boundaries were occupied by structural maintenance of chromosomes (SMC1) cohesin complex and CCCTC-binding factor (CTCF), and the KSHV transactivator was recruited to those sites during reactivation. Triggering KSHV gene expression decreased prewired genomic loops within the regulatory unit, while contacts extending outside of regulatory borders increased, leading to formation of a larger regulatory unit with a shift from repressive to active compartments (B to A). The 3D genomic structural model proposes that the immediate early promoter region is localized on the periphery of the 3D viral genome during latency, while highly inducible noncoding RNA regions moved toward the inner space of the structure, resembling the configuration of a "bird cage" during reactivation. The compartment-like properties of viral episomal chromatin structure and its reorganization during the transition from latency may help facilitate viral gene transcription. IMPORTANCE The 3D architecture of chromatin allows for efficient arrangement, expression, and replication of genetic material. The genomes of all organisms studied to date have been found to be organized through some form of tiered domain structures. However, the architectural framework of the genomes of large double-stranded DNA viruses such as the herpesvirus family has not been reported. Prior studies with Kaposi's sarcoma-associated herpesvirus (KSHV) have indicated that the viral chromatin shares many biological properties exhibited by the host cell genome, essentially behaving as a mini human chromosome. Thus, we hypothesized that the KSHV genome may be organized in a similar manner. In this report, we describe the domain structure of the latent and lytic KSHV genome at 500 bp resolution and present a 3D genomic structural model for KSHV under each condition. These results add new insights into the complex regulation of the viral life cycle.
Keywords: CTCF; Capture Hi-C; KSHV; ORF50; TAD; epigenetics; genome organization; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10:e1003847. 10.1371/journal.ppat.1003847. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI155515/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI167663/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI167663/AI/NIAID NIH HHS/United States
- CA225266/HHS | NIH | National Cancer Institute (NCI)
- R21 AI155515/AI/NIAID NIH HHS/United States
- DE025985/HHS | NIH | National Institute of Dental and Craniofacial Research (NIDCR)
- P30 CA093373/CA/NCI NIH HHS/United States
- R21 AI147207/AI/NIAID NIH HHS/United States
- R01 CA232845/CA/NCI NIH HHS/United States
- AI147207/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- CA232845/HHS | NIH | National Cancer Institute (NCI)
- R01 CA225266/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

