Excreted Antibiotics May Be Key to Emergence of Increasingly Efficient Antibiotic Resistance in Food Animal Production
- PMID: 35867586
- PMCID: PMC9361830
- DOI: 10.1128/aem.00791-22
Excreted Antibiotics May Be Key to Emergence of Increasingly Efficient Antibiotic Resistance in Food Animal Production
Abstract
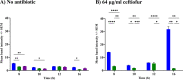
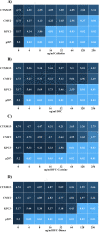
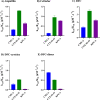
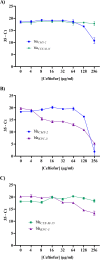
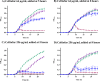
At a time when antibiotic resistance is seemingly ubiquitous worldwide, understanding the mechanisms responsible for successful emergence of new resistance genes may provide insights into the persistence and pathways of dissemination for antibiotic-resistant organisms in general. For example, Escherichia coli strains harboring a class A β-lactamase-encoding gene (blaCTX-M-15) appear to be displacing strains that harbor a class C β-lactamase gene (blaCMY-2) in Washington State dairy cattle. We cloned these genes with native promoters into low-copy-number plasmids that were then transformed into isogenic strains of E. coli, and growth curves were generated for two commonly administered antibiotics (ampicillin and ceftiofur). Both strains met the definition of resistance for ampicillin (≥32 μg/mL) and ceftiofur (≥16 μg/mL). Growth of the CMY-2-producing strain was compromised at 1,000 μg/mL ampicillin, whereas the CTX-M-15-producing strain was not inhibited in the presence of 3,000 μg/mL ampicillin or with most concentrations of ceftiofur, although there were mixed outcomes with ceftiofur metabolites. Consequently, in the absence of competing genes, E. coli harboring either gene would experience a selective advantage if exposed to these antibiotics. Successful emergence of CTX-M-15-producing strains where CMY-2-producing strains are already established, however, requires high concentrations of antibiotics that can only be found in the urine of treated animals (e.g., >2,000 μg/mL for ampicillin, based on literature). This ex vivo selection pressure may be important for the emergence of new and more efficient antibiotic resistance genes and likely for persistence of antibiotic-resistant bacteria in food animal populations. IMPORTANCE We studied the relative fitness benefits of a cephalosporin resistance enzyme (CTX-M-15) that is displacing a similar enzyme (CMY-2), which is extant in E. coli from dairy cattle in Washington State. In vitro experiments demonstrated that CTX-M-15 provides a significant fitness advantage, but only in the presence of very high concentrations of antibiotic that are only found when the antibiotic ampicillin, and to a lesser extent ceftiofur, is excreted in urine from treated animals. As such, the increasing prevalence of bacteria with blaCTX-M-15 is likely occurring ex vivo. Interventions should focus on controlling waste from treated animals and, when possible, selecting antibiotics that are less likely to impact the proximal environment of treated animals.
Keywords: antibiotic; antibiotic resistance; antimicrobial; blaCMY-2; blaCTX-M-15; blaKPC-3; competition; fitness; hydrolysis; resistance; β-lactamase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. 10.1016/S1473-3099(15)00424-7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

