The ZCCHC14/TENT4 complex is required for hepatitis A virus RNA synthesis
- PMID: 35867748
- PMCID: PMC9282228
- DOI: 10.1073/pnas.2204511119
The ZCCHC14/TENT4 complex is required for hepatitis A virus RNA synthesis
Abstract
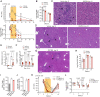
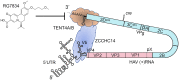
Despite excellent vaccines, resurgent outbreaks of hepatitis A have caused thousands of hospitalizations and hundreds of deaths within the United States in recent years. There is no effective antiviral therapy for hepatitis A, and many aspects of the hepatitis A virus (HAV) replication cycle remain to be elucidated. Replication requires the zinc finger protein ZCCHC14 and noncanonical TENT4 poly(A) polymerases with which it associates, but the underlying mechanism is unknown. Here, we show that ZCCHC14 and TENT4A/B are required for viral RNA synthesis following translation of the viral genome in infected cells. Cross-linking immunoprecipitation sequencing (CLIP-seq) experiments revealed that ZCCHC14 binds a small stem-loop in the HAV 5' untranslated RNA possessing a Smaug recognition-like pentaloop to which it recruits TENT4. TENT4 polymerases lengthen and stabilize the 3' poly(A) tails of some cellular and viral mRNAs, but the chemical inhibition of TENT4A/B with the dihydroquinolizinone RG7834 had no impact on the length of the HAV 3' poly(A) tail, stability of HAV RNA, or cap-independent translation of the viral genome. By contrast, RG7834 inhibited the incorporation of 5-ethynyl uridine into nascent HAV RNA, indicating that TENT4A/B function in viral RNA synthesis. Consistent with potent in vitro antiviral activity against HAV (IC50 6.11 nM), orally administered RG7834 completely blocked HAV infection in Ifnar1-/- mice, and sharply reduced serum alanine aminotransferase activities, hepatocyte apoptosis, and intrahepatic inflammatory cell infiltrates in mice with acute hepatitis A. These results reveal requirements for ZCCHC14-TENT4A/B in hepatovirus RNA synthesis, and suggest that TENT4A/B inhibitors may be useful for preventing or treating hepatitis A in humans.
Keywords: RNA-binding protein; animal model; antiviral therapy; hepatitis A; picornavirus.
Conflict of interest statement
Y.L. and S.M.L. are co-inventors on a patent application related to DHQ-E-OH. The other authors declare no competing interest.
Figures



References
-
- Centers for Disease Control and Prevention, Hepatitis Outbreaks. (2021). https://www.cdc.gov/hepatitis/outbreaks/2017March-HepatitisA.htm
-
- Lemon S. M., Ott J. J., Van Damme P., Shouval D., Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 68, 167–184 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-AI138337/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI143894/AI/NIAID NIH HHS/United States
- R01-AI143894/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI138337/AI/NIAID NIH HHS/United States
- R01 AI131685/AI/NIAID NIH HHS/United States
- R01-AI150095/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI150095/AI/NIAID NIH HHS/United States
- R01-AI103083/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI103083/AI/NIAID NIH HHS/United States
- R01-AI131685/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P30 CA016086/CA/NCI NIH HHS/United States
- R21 AI163606/AI/NIAID NIH HHS/United States
- R21-AI163606/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

