Spatiotemporal gating of Stat nuclear influx by Drosophila Npas4 in collective cell migration
- PMID: 35867785
- PMCID: PMC9307255
- DOI: 10.1126/sciadv.abm2411
Spatiotemporal gating of Stat nuclear influx by Drosophila Npas4 in collective cell migration
Abstract
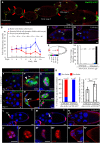
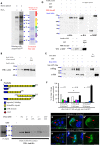
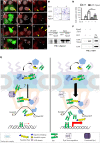
Collective migration is important to embryonic development and cancer metastasis, but migratory and nonmigratory cell fate discrimination by differential activity of signal pathways remains elusive. In Drosophila oogenesis, Jak/Stat signaling patterns the epithelial cell fates in early egg chambers but later renders motility to clustered border cells. How Jak/Stat signal spatiotemporally switches static epithelia to motile cells is largely unknown. We report that a nuclear protein, Dysfusion, resides on the inner nuclear membrane and interacts with importin α/β and Nup153 to modulate Jak/Stat signal by attenuating Stat nuclear import. Dysfusion is ubiquitously expressed in oogenesis but specifically down-regulated in border cells when migrating. Increase of nuclear Stat by Dysfusion down-regulation triggers invasive cell behavior and maintains persistent motility. Mammalian homolog of Dysfusion (NPAS4) also negatively regulates the nuclear accumulation of STAT3 and cancer cell migration. Thus, our finding demonstrates that Dysfusion-dependent gating mechanism is conserved and may serve as a therapeutic target for Stat-mediated cancer metastasis.
Figures




Similar articles
-
Socs36E limits STAT signaling via Cullin2 and a SOCS-box independent mechanism in the Drosophila egg chamber.Mech Dev. 2015 Nov;138 Pt 3:313-27. doi: 10.1016/j.mod.2015.08.003. Epub 2015 Aug 13. Mech Dev. 2015. PMID: 26277564
-
Identification of Novel Regulators of the JAK/STAT Signaling Pathway that Control Border Cell Migration in the Drosophila Ovary.G3 (Bethesda). 2016 Jul 7;6(7):1991-2002. doi: 10.1534/g3.116.028100. G3 (Bethesda). 2016. PMID: 27175018 Free PMC article.
-
The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis.Development. 2002 Dec;129(23):5437-47. doi: 10.1242/dev.00116. Development. 2002. PMID: 12403714
-
Drosophila Jak/STAT Signaling: Regulation and Relevance in Human Cancer and Metastasis.Int J Mol Sci. 2018 Dec 14;19(12):4056. doi: 10.3390/ijms19124056. Int J Mol Sci. 2018. PMID: 30558204 Free PMC article. Review.
-
Organogenesis and tumorigenesis: insight from the JAK/STAT pathway in the Drosophila eye.Dev Dyn. 2010 Oct;239(10):2522-33. doi: 10.1002/dvdy.22394. Dev Dyn. 2010. PMID: 20737505 Free PMC article. Review.
Cited by
-
Detecting Native Protein-Protein Interactions by APEX2 Proximity Labeling in Drosophila Tissues.Bio Protoc. 2024 Oct 20;14(20):e5090. doi: 10.21769/BioProtoc.5090. eCollection 2024 Oct 20. Bio Protoc. 2024. PMID: 39512889 Free PMC article.
-
Nuclear lamin facilitates collective border cell invasion into confined spaces in vivo.J Cell Biol. 2023 Nov 6;222(11):e202212101. doi: 10.1083/jcb.202212101. Epub 2023 Sep 11. J Cell Biol. 2023. PMID: 37695420 Free PMC article.
References
-
- Hogarth C., Itman C., Jans D. A., Loveland K. L., Regulated nucleocytoplasmic transport in spermatogenesis: A driver of cellular differentiation? Bioessays 27, 1011–1025 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

