Synthetic chromosomes, genomes, viruses, and cells
- PMID: 35868275
- PMCID: PMC9347161
- DOI: 10.1016/j.cell.2022.06.046
Synthetic chromosomes, genomes, viruses, and cells
Abstract
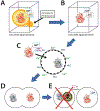
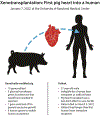
Synthetic genomics is the construction of viruses, bacteria, and eukaryotic cells with synthetic genomes. It involves two basic processes: synthesis of complete genomes or chromosomes and booting up of those synthetic nucleic acids to make viruses or living cells. The first synthetic genomics efforts resulted in the construction of viruses. This led to a revolution in viral reverse genetics and improvements in vaccine design and manufacture. The first bacterium with a synthetic genome led to construction of a minimal bacterial cell and recoded Escherichia coli strains able to incorporate multiple non-standard amino acids in proteins and resistant to phage infection. Further advances led to a yeast strain with a synthetic genome and new approaches for animal and plant artificial chromosomes. On the horizon there are dramatic advances in DNA synthesis that will enable extraordinary new opportunities in medicine, industry, agriculture, and research.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The JCVI and J.C.V. have shares in Synthetic Genomics, Inc. (now named Viridos). J.C.V. is on the board of directors of Avery Digital Data and owns less than 1% of the company stock. J.I.G. is an advisor for Avery Digital Data. C.A.H. and S.V. declare no competing interests. The authors are inventors on the following synthetic genomics-related patents.
Figures



References
-
- Addressing Biosecurity Concerns Related to the Synthesis of Select Agents. (2006). https://osp.od.nih.gov/wp-content/uploads/2013/06/Final_NSABB_Report_on_....
-
- Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, and Denison MR (2008). Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105, 19944–19949. 10.1073/pnas.0808116105. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

