A comparison of high-throughput SARS-CoV-2 sequencing methods from nasopharyngeal samples
- PMID: 35869099
- PMCID: PMC9306416
- DOI: 10.1038/s41598-022-16549-w
A comparison of high-throughput SARS-CoV-2 sequencing methods from nasopharyngeal samples
Abstract
The COVID-19 pandemic caused by the new Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) continues to threaten public health and burden healthcare systems worldwide. Whole SARS-CoV-2 genome sequencing has become essential for epidemiological monitoring and identification of new variants, which could represent a risk of increased transmissibility, virulence, or resistance to vaccines or treatment. Different next-generation sequencing approaches are used in SARS-CoV-2 sequencing, although with different ability to provide whole genome coverage without gaps and to reliably detect new variants. In this study, we compared the performance of three target enrichment methods (two multiplex amplification methods and one hybridization capture) using nasopharyngeal swabs from infected individuals. We applied these target enrichment methods to the same set of nasopharyngeal samples (N = 93) in high-throughput mode. SARS-CoV-2 genome was obtained using short-read next-generation sequencing. We observed that each method has some advantages, such as high mapping rate (CleanPlex and COVIDSeq) or absence of systematic variant calling error (SureSelect) as well as their limitations such as suboptimal uniformity of coverage (CleanPlex), high cost (SureSelect) or supply shortages (COVIDSeq). Nevertheless, each of the three target enrichment kits tested in this study yielded acceptable results of whole SARS-CoV-2 genome sequencing and either of them can therefore be used in prospective programs of genomic surveillance of SARS-CoV-2. Genomic surveillance will be crucial to overcoming the ongoing pandemic of COVID-19, despite its successive waves and continually emerging variants.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
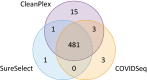
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

