Investigation of rumen long noncoding RNA before and after weaning in cattle
- PMID: 35869425
- PMCID: PMC9308236
- DOI: 10.1186/s12864-022-08758-4
Investigation of rumen long noncoding RNA before and after weaning in cattle
Abstract
Background: This study aimed to identify long non-coding RNA (lncRNA) from the rumen tissue in dairy cattle, explore their features including expression and conservation levels, and reveal potential links between lncRNA and complex traits that may indicate important functional impacts of rumen lncRNA during the transition to the weaning period.
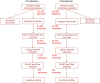
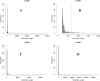
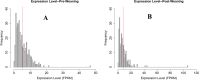
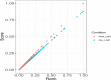
Results: A total of six cattle rumen samples were taken with three replicates from before and after weaning periods, respectively. Total RNAs were extracted and sequenced with lncRNA discovered based on size, coding potential, sequence homology, and known protein domains. As a result, 404 and 234 rumen lncRNAs were identified before and after weaning, respectively. However, only nine of them were shared under two conditions, with 395 lncRNAs found only in pre-weaning tissues and 225 only in post-weaning samples. Interestingly, none of the nine common lncRNAs were differentially expressed between the two weaning conditions. LncRNA averaged shorter length, lower expression, and lower conservation scores than the genome overall, which is consistent with general lncRNA characteristics. By integrating rumen lncRNA before and after weaning with large-scale GWAS results in cattle, we reported significant enrichment of both pre- and after-weaning lncRNA with traits of economic importance including production, reproduction, health, and body conformation phenotypes.
Conclusions: The majority of rumen lncRNAs are uniquely expressed in one of the two weaning conditions, indicating a functional role of lncRNA in rumen development and transition of weaning. Notably, both pre- and post-weaning lncRNA showed significant enrichment with a variety of complex traits in dairy cattle, suggesting the importance of rumen lncRNA for cattle performance in the adult stage. These relationships should be further investigated to better understand the specific roles lncRNAs are playing in rumen development and cow performance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species.BMC Genomics. 2018 Sep 18;19(1):684. doi: 10.1186/s12864-018-5037-7. BMC Genomics. 2018. PMID: 30227846 Free PMC article.
-
Genome-wide identification and characterization of novel long non-coding RNA in Ruminal tissue affected with sub-acute Ruminal acidosis from Holstein cattle.Vet Res Commun. 2020 Feb;44(1):19-27. doi: 10.1007/s11259-020-09769-w. Epub 2020 Feb 11. Vet Res Commun. 2020. PMID: 32043213
-
Genome-wide detection and sequence conservation analysis of long non-coding RNA during hair follicle cycle of yak.BMC Genomics. 2020 Oct 1;21(1):681. doi: 10.1186/s12864-020-07082-z. BMC Genomics. 2020. PMID: 32998696 Free PMC article.
-
Utilization of digital differential display to identify differentially expressed genes related to rumen development.Anim Sci J. 2016 Apr;87(4):584-90. doi: 10.1111/asj.12448. Epub 2015 Sep 21. Anim Sci J. 2016. PMID: 26388291 Review.
-
On the classification of long non-coding RNAs.RNA Biol. 2013 Jun;10(6):925-33. doi: 10.4161/rna.24604. Epub 2013 Apr 15. RNA Biol. 2013. PMID: 23696037 Free PMC article. Review.
Cited by
-
Long non-coding RNA (LncRNA) and epigenetic factors: their role in regulating the adipocytes in bovine.Front Genet. 2024 Oct 3;15:1405588. doi: 10.3389/fgene.2024.1405588. eCollection 2024. Front Genet. 2024. PMID: 39421300 Free PMC article. Review.
-
Enhanced bovine genome annotation through integration of transcriptomics and epi-transcriptomics datasets facilitates genomic biology.Gigascience. 2024 Jan 2;13:giae019. doi: 10.1093/gigascience/giae019. Gigascience. 2024. PMID: 38626724 Free PMC article.
References
-
- Jia X, He Y, Chen S-Y, Wang J, Hu S, Lai S-J. Genome-wide identification and characterisation of long non-coding RNAs in two Chinese cattle breeds. Italian Journal of Animal Science. 2020;19(1):383–391. doi: 10.1080/1828051X.2020.1735266. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

