Proteomics data analysis using multiple statistical approaches identified proteins and metabolic networks associated with sucrose accumulation in sugarcane
- PMID: 35869434
- PMCID: PMC9308345
- DOI: 10.1186/s12864-022-08768-2
Proteomics data analysis using multiple statistical approaches identified proteins and metabolic networks associated with sucrose accumulation in sugarcane
Abstract
Background: Sugarcane is the most important sugar crop, contributing > 80% of global sugar production. High sucrose content is a key target of sugarcane breeding, yet sucrose improvement in sugarcane remains extremely slow for decades. Molecular breeding has the potential to break through the genetic bottleneck of sucrose improvement. Dissecting the molecular mechanism(s) and identifying the key genetic elements controlling sucrose accumulation will accelerate sucrose improvement by molecular breeding. In our previous work, a proteomics dataset based on 12 independent samples from high- and low-sugar genotypes treated with ethephon or water was established. However, in that study, employing conventional analysis, only 25 proteins involved in sugar metabolism were identified .
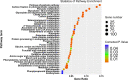
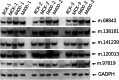
Results: In this work, the proteomics dataset used in our previous study was reanalyzed by three different statistical approaches, which include a logistic marginal regression, a penalized multiple logistic regression named Elastic net, as well as a Bayesian multiple logistic regression method named Stochastic search variable selection (SSVS) to identify more sugar metabolism-associated proteins. A total of 507 differentially abundant proteins (DAPs) were identified from this dataset, with 5 of them were validated by western blot. Among the DAPs, 49 proteins were found to participate in sugar metabolism-related processes including photosynthesis, carbon fixation as well as carbon, amino sugar, nucleotide sugar, starch and sucrose metabolism. Based on our studies, a putative network of key proteins regulating sucrose accumulation in sugarcane is proposed, with glucose-6-phosphate isomerase, 2-phospho-D-glycerate hydrolyase, malate dehydrogenase and phospho-glycerate kinase, as hub proteins.
Conclusions: The sugar metabolism-related proteins identified in this work are potential candidates for sucrose improvement by molecular breeding. Further, this work provides an alternative solution for omics data processing.
Keywords: Differentially abundant protein; Proteomics; Statistical approach; Sucrose accumulation; Sugarcane.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the Lasso. Ann Stat. 2006;34:1436–1462.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

