PM2.5 promotes NSCLC carcinogenesis through translationally and transcriptionally activating DLAT-mediated glycolysis reprograming
- PMID: 35869499
- PMCID: PMC9308224
- DOI: 10.1186/s13046-022-02437-8
PM2.5 promotes NSCLC carcinogenesis through translationally and transcriptionally activating DLAT-mediated glycolysis reprograming
Abstract
Background: Airborne fine particulate matter (PM2.5) has been associated with lung cancer development and progression in never smokers. However, the molecular mechanisms underlying PM2.5-induced lung cancer remain largely unknown. The aim of this study was to explore the mechanisms by which PM2.5 regulated the carcinogenesis of non-small cell lung cancer (NSCLC).
Methods: Paralleled ribosome sequencing (Ribo-seq) and RNA sequencing (RNA-seq) were performed to identify PM2.5-associated genes for further study. Quantitative real time-PCR (qRT-PCR), Western blot, and immunohistochemistry (IHC) were used to determine mRNA and protein expression levels in tissues and cells. The biological roles of PM2.5 and PM2.5-dysregulated gene were assessed by gain- and loss-of-function experiments, biochemical analyses, and Seahorse XF glycolysis stress assays. Human tissue microarray analysis and 18F-FDG PET/CT scans in patients with NSCLC were used to verify the experimental findings. Polysome fractionation experiments, chromatin immunoprecipitation (ChIP), and dual-luciferase reporter assay were implemented to explore the molecular mechanisms.
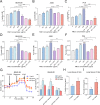
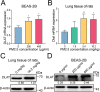
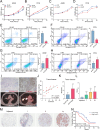
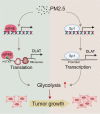
Results: We found that PM2.5 induced a translation shift towards glycolysis pathway genes and increased glycolysis metabolism, as evidenced by increased L-lactate and pyruvate concentrations or higher extracellular acidification rate (ECAR) in vitro and in vivo. Particularly, PM2.5 enhanced the expression of glycolytic gene DLAT, which promoted glycolysis but suppressed acetyl-CoA production and enhanced the malignancy of NSCLC cells. Clinically, high expression of DLAT was positively associated with tumor size, poorer prognosis, and SUVmax values of 18F-FDG-PET/CT scans in patients with NSCLC. Mechanistically, PM2.5 activated eIF4E, consequently up-regulating the expression level of DLAT in polysomes. PM2.5 also stimulated transcription factor Sp1, which further augmented transcription activity of DLAT promoter.
Conclusions: This study demonstrated that PM2.5-activated overexpression of DLAT and enhancement in glycolysis metabolism contributed to the tumorigenesis of NSCLC, suggesting that DLAT-associated pathway may be a therapeutic target for NSCLC.
Keywords: DLAT; Glycolysis reprograming; Non-small cell lung cancer (NSCLC); PM2.5; Sp1; Transcription; Translation; eIF4E.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- 41977372/National Natural Science Foundation of China
- 81903412/National Natural Science Foundation of China
- 117-00004112/Natural Science Foundation of Guangdong Province
- 2022A1515012033/Natural Science Foundation of Guangdong Province
- JCYJ20190806154210829/Science, Technology and Innovation Commission of Shenzhen Municipality
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

