Neuroscience Cloud Analysis As a Service: An open-source platform for scalable, reproducible data analysis
- PMID: 35870448
- PMCID: PMC9464703
- DOI: 10.1016/j.neuron.2022.06.018
Neuroscience Cloud Analysis As a Service: An open-source platform for scalable, reproducible data analysis
Abstract
A key aspect of neuroscience research is the development of powerful, general-purpose data analyses that process large datasets. Unfortunately, modern data analyses have a hidden dependence upon complex computing infrastructure (e.g., software and hardware), which acts as an unaddressed deterrent to analysis users. Although existing analyses are increasingly shared as open-source software, the infrastructure and knowledge needed to deploy these analyses efficiently still pose significant barriers to use. In this work, we develop Neuroscience Cloud Analysis As a Service (NeuroCAAS): a fully automated open-source analysis platform offering automatic infrastructure reproducibility for any data analysis. We show how NeuroCAAS supports the design of simpler, more powerful data analyses and that many popular data analysis tools offered through NeuroCAAS outperform counterparts on typical infrastructure. Pairing rigorous infrastructure management with cloud resources, NeuroCAAS dramatically accelerates the dissemination and use of new data analyses for neuroscientific discovery.
Keywords: cloud compute; data analysis; ensembling; infrastructure-as-code; markerless tracking; open source; widefield calcium imaging.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



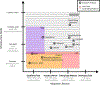
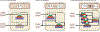
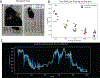

Comment in
-
Neuroscience data analysis in the cloud.Nat Methods. 2022 Sep;19(9):1034. doi: 10.1038/s41592-022-01613-0. Nat Methods. 2022. PMID: 36068316 No abstract available.
References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M et al. (2016), Tensorflow: A system for large-scale machine learning, in ‘12th Symposium on Operating Systems Design and Implementation (OSDI 16)’, pp. 265–283.
-
- Aguiar A, Díaz J, Almaraz R, Pérez J and Garbajosa J (2018), DevOps in Practice – An Exploratory Case Study, in ‘Proceedings of the 19th International Conference on Agile Software Development: Companion, XP ‘18’, pp. 1–3.
-
- Amstutz P, Crusoe MR, Tijanic N. s., Chapman B, Chilton J, Heuer M, Kartashov A, Kern J, Leehr D, Menager H, Nedeljkovich M, Scales M, Soiland-Reyes S and Stojanovic L (2016), ‘Common Workflow Language, v1.0’. 10.6084/m9.figshare.3115156.v2 - DOI
-
- Avesani P, McPherson B, Hayashi S, Caiafa CF, Henschel R, Garyfallidis E, Kitchell L, Bullock D, Patterson A, Olivetti E et al. (2019), ‘The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services’, Scientific data 6(1), 1–13. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

