The flax genome reveals orbitide diversity
- PMID: 35870878
- PMCID: PMC9308333
- DOI: 10.1186/s12864-022-08735-x
The flax genome reveals orbitide diversity
Abstract
Background: Ribosomally-synthesized cyclic peptides are widely found in plants and exhibit useful bioactivities for humans. The identification of cyclic peptide sequences and their precursor proteins is facilitated by the growing number of sequenced genomes. While previous research largely focused on the chemical diversity of these peptides across various species, there is little attention to a broader range of potential peptides that are not chemically identified.
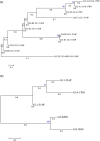
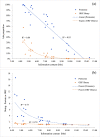
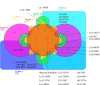
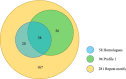
Results: A pioneering study was initiated to explore the genetic diversity of linusorbs, a group of cyclic peptides uniquely occurring in cultivated flax (Linum usitatissimum). Phylogenetic analysis clustered the 5 known linusorb precursor proteins into two clades and one singleton. Preliminary tBLASTn search of the published flax genome using the whole protein sequence as query could only retrieve its homologues within the same clade. This limitation was overcome using a profile-based mining strategy. After genome reannotation, a hidden Markov Model (HMM)-based approach identified 58 repeats homologous to the linusorb-embedded repeats in 8 novel proteins, implying that they share common ancestry with the linusorb-embedded repeats. Subsequently, we developed a customized profile composed of a random linusorb-like domain (LLD) flanked by 5 conserved sites and used it for string search of the proteome, which extracted 281 LLD-containing repeats (LLDRs) in 25 proteins. Comparative analysis of different repeat categories suggested that the 5 conserved flanking sites among the non-homologous repeats have undergone convergent evolution driven by functional selection.
Conclusions: The profile-based mining approach is suitable for analyzing repetitive sequences. The 25 LLDR proteins identified herein represent the potential diversity of cyclic peptides within the flax genome and lay a foundation for further studies on the functions and evolution of these protein tandem repeats.
Keywords: Diversity; Linum usitatissimum; Mining; Orbitide; Protein tandem repeats.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Novel flax orbitide derived from genetic deletion.BMC Plant Biol. 2018 May 21;18(1):90. doi: 10.1186/s12870-018-1303-8. BMC Plant Biol. 2018. PMID: 29783946 Free PMC article.
-
[Biological activity and prospects for food application of cyclic peptides of flax (Linum usitatissimum)].Vopr Pitan. 2024;93(6):37-48. doi: 10.33029/0042-8833-2024-93-6-37-48. Epub 2024 Dec 5. Vopr Pitan. 2024. PMID: 39887163 Russian.
-
Characterization of repeated DNA sequences in genomes of blue-flowered flax.BMC Evol Biol. 2019 Feb 26;19(Suppl 1):49. doi: 10.1186/s12862-019-1375-6. BMC Evol Biol. 2019. PMID: 30813893 Free PMC article.
-
Mechanisms and control of rapid genomic changes in flax.Ann Bot. 2005 Jan;95(1):201-6. doi: 10.1093/aob/mci013. Ann Bot. 2005. PMID: 15596467 Free PMC article. Review.
-
A Review on Pharmacological and Clinical Aspects of Linum usitatissimum L.Curr Drug Discov Technol. 2019;16(2):148-158. doi: 10.2174/1570163815666180521101136. Curr Drug Discov Technol. 2019. PMID: 29779483 Review.
Cited by
-
Plant peptides - redefining an area of ribosomally synthesized and post-translationally modified peptides.Nat Prod Rep. 2024 Jul 17;41(7):1020-1059. doi: 10.1039/d3np00042g. Nat Prod Rep. 2024. PMID: 38411572 Free PMC article. Review.
-
Large-scale transcriptome mining enables macrocyclic diversification and improved bioactivity of the stephanotic acid scaffold.Nat Commun. 2025 May 6;16(1):4198. doi: 10.1038/s41467-025-59428-4. Nat Commun. 2025. PMID: 40328797 Free PMC article.
-
Identification of a key peptide cyclase for novel cyclic peptide discovery in Pseudostellaria heterophylla.Plant Commun. 2025 May 12;6(5):101315. doi: 10.1016/j.xplc.2025.101315. Epub 2025 Mar 13. Plant Commun. 2025. PMID: 40083160 Free PMC article.
References
-
- Shim YY, Song Z, Jadhav PD, Reaney MJT. Orbitides from flaxseed (Linum usitatissimum L.): a comprehensive review. Trends Food Sci Technol. 2019;93:197–211. doi: 10.1016/j.tifs.2019.09.007. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

