Analysis of gastric microbiome reveals three distinctive microbial communities associated with the occurrence of gastric cancer
- PMID: 35870901
- PMCID: PMC9308235
- DOI: 10.1186/s12866-022-02594-y
Analysis of gastric microbiome reveals three distinctive microbial communities associated with the occurrence of gastric cancer
Abstract
Background: Gastric microbial dysbiosis were reported to be associated with gastric cancer (GC). This study aimed to explore the variation, diversity, and composition patterns of gastric bacteria in stages of gastric carcinogenesis based on the published datasets.
Methods: We conducted a gastric microbial analysis using 10 public datasets based on 16S rRNA sequencing, including 1270 gastric biopsies of 109 health control, 183 superficial gastritis (SG), 135 atrophic gastritis (AG), 124 intestinal metaplasia (IM), 94 intraepithelial neoplasia (IN), 344 GC, and 281 adjacent normal tissues. And QIIME2-pipeline, DESeq2, NetMoss2, vegan, igraph, and RandomForest were used for the data processing and analysis.
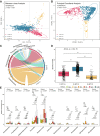
Results: We identified three gastric microbial communities among all the gastric tissues. The first community (designate as GT-H) was featured by the high abundance of Helicobacter. The other two microbial communities, namely GT-F, and GT-P, were featured by the enrichment of phylum Firmicutes and Proteobacteria, respectively. The distribution of GC-associated bacteria, such as Fusobacterium, Peptostreptococcus, Streptococcus, and Veillonella were enriched in tumor tissues, and mainly distributed in GT-F type microbial communities. Compared with SG, AG, and IM, the bacterial diversity in GC was significantly reduced. And the strength of microbial interaction networks was initially increased in IM but gradually decreased from IN to GC. In addition, Randomforest models constructed in in GT-H and GT-F microbial communities showed excellent performance in distinguishing GC from SG and precancerous stages, with varied donated bacteria.
Conclusions: This study identified three types of gastric microbiome with different patterns of composition which helps to clarify the potential key bacteria in the development of gastric carcinogenesis.
Keywords: Bacterial community; Gastric cancer; Microbiota; Predictive model.
© 2022. The Author(s).
Conflict of interest statement
I declare that the authors have no competing interests as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

