Comparing dormancy in two distantly related tunicates reveals morphological, molecular, and ecological convergences and repeated co-option
- PMID: 35871255
- PMCID: PMC9308810
- DOI: 10.1038/s41598-022-16656-8
Comparing dormancy in two distantly related tunicates reveals morphological, molecular, and ecological convergences and repeated co-option
Abstract
Many asexually-propagating marine invertebrates can survive extreme environmental conditions by developing dormant structures, i.e., morphologically simplified bodies that retain the capacity to completely regenerate a functional adult when conditions return to normal. Here, we examine the environmental, morphological, and molecular characteristics of dormancy in two distantly related clonal tunicate species: Polyandrocarpa zorritensis and Clavelina lepadiformis. In both species, we report that the dormant structures are able to withstand harsher temperature and salinity conditions compared to the adults. The dormant structures are the dominant forms these species employ to survive adverse conditions when the zooids themselves cannot survive. While previous work shows C. lepadiformis dormant stage is present in winters in the Atlantic Ocean and summers in the Mediterranean, this study is the first to show a year-round presence of P. zorritensis dormant forms in NW Italy, even in the late winter when all zooids have disappeared. By finely controlling the entry and exit of dormancy in laboratory-reared individuals, we were able to select and characterize the morphology of dormant structures associated with their transcriptome dynamics. In both species, we identified putative stem and nutritive cells in structures that resemble the earliest stages of asexual propagation. By characterizing gene expression during dormancy and regeneration into the adult body plan (i.e., germination), we observed that genes which control dormancy and environmental sensing in other metazoans, notably HIF-α and insulin signaling genes, are also expressed in tunicate dormancy. Germination-related genes in these two species, such as the retinoic acid pathway, are also found in other unrelated clonal tunicates during asexual development. These results are suggestive of repeated co-option of conserved eco-physiological and regeneration programs for the origin of novel dormancy-germination processes across distantly related animal taxa.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



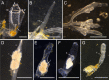



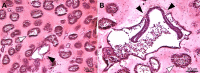



Similar articles
-
A transcriptomic examination of encased rotifer embryos reveals the developmental trajectory leading to long-term dormancy; are they "animal seeds"?BMC Genomics. 2024 Jan 27;25(1):119. doi: 10.1186/s12864-024-09961-1. BMC Genomics. 2024. PMID: 38281016 Free PMC article.
-
Dormancy cycling: translation-related transcripts are the main difference between dormant and non-dormant seeds in the field.Plant J. 2020 Apr;102(2):327-339. doi: 10.1111/tpj.14626. Epub 2020 Feb 5. Plant J. 2020. PMID: 31785171 Free PMC article.
-
Differentially expressed genes during the imbibition of dormant and after-ripened seeds - a reverse genetics approach.BMC Plant Biol. 2017 Sep 11;17(1):151. doi: 10.1186/s12870-017-1098-z. BMC Plant Biol. 2017. PMID: 28893189 Free PMC article.
-
Dormancy and germination: How does the crop seed decide?Plant Biol (Stuttg). 2015 Nov;17(6):1104-12. doi: 10.1111/plb.12356. Epub 2015 Jul 14. Plant Biol (Stuttg). 2015. PMID: 26095078 Review.
-
A graphical method for identifying the six types of non-deep physiological dormancy in seeds.Plant Biol (Stuttg). 2017 Sep;19(5):673-682. doi: 10.1111/plb.12590. Epub 2017 Jun 28. Plant Biol (Stuttg). 2017. PMID: 28612366 Review.
Cited by
-
Uncovering developmental diversity in the field.Development. 2024 Oct 15;151(20):dev203084. doi: 10.1242/dev.203084. Epub 2024 Aug 19. Development. 2024. PMID: 39158021 Free PMC article.
-
Genomic Resources and Annotations for a Colonial Ascidian, the Light-Bulb Sea Squirt Clavelina lepadiformis.Genome Biol Evol. 2024 Mar 2;16(3):evae038. doi: 10.1093/gbe/evae038. Genome Biol Evol. 2024. PMID: 38441487 Free PMC article.
References
-
- Hand, S.C. Metabolic dormancy in aquatic invertebrates. In Advances in Comparative and Environmental Physiology, Vol. 8 (ed. Gilles, R.) 1–50. 10.1007/978-3-642-75900-0_1 (1991).
-
- Cáceres CE. Dormancy in Invertebrates. Invertebr. Biol. 1997;116(4):371–383. doi: 10.2307/3226870. - DOI
-
- Wilsterman K, Ballinger MA, Williams CM. A unifying, eco-physiological framework for animal dormancy. Funct. Ecol. 2021;35:11–31. doi: 10.1111/1365-2435.13718. - DOI
-
- Bertolani R, Guidetti R, Altiero T, Nelson DR, Rebecchi L. Dormancy in Freshwater Tardigrades. In: Alekseev V, Pinel-Alloul B, editors. Dormancy in Aquatic Organisms. Theory, Human Use and Modeling. Monographiae Biologicae. Cham: Springer; 2019.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

