SUPT3H-less SAGA coactivator can assemble and function without significantly perturbing RNA polymerase II transcription in mammalian cells
- PMID: 35871303
- PMCID: PMC9371916
- DOI: 10.1093/nar/gkac637
SUPT3H-less SAGA coactivator can assemble and function without significantly perturbing RNA polymerase II transcription in mammalian cells
Abstract
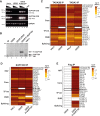
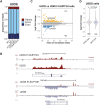
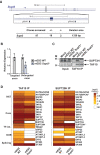
Coactivator complexes regulate chromatin accessibility and transcription. SAGA (Spt-Ada-Gcn5 Acetyltransferase) is an evolutionary conserved coactivator complex. The core module scaffolds the entire SAGA complex and adopts a histone octamer-like structure, which consists of six histone-fold domain (HFD)-containing proteins forming three histone-fold (HF) pairs, to which the double HFD-containing SUPT3H adds one HF pair. Spt3, the yeast ortholog of SUPT3H, interacts genetically and biochemically with the TATA binding protein (TBP) and contributes to global RNA polymerase II (Pol II) transcription. Here we demonstrate that (i) SAGA purified from human U2OS or mouse embryonic stem cells (mESC) can assemble without SUPT3H, (ii) SUPT3H is not essential for mESC survival, but required for their growth and self-renewal, and (iii) the loss of SUPT3H from mammalian cells affects the transcription of only a specific subset of genes. Accordingly, in the absence of SUPT3H no major change in TBP accumulation at gene promoters was observed. Thus, SUPT3H is not required for the assembly of SAGA, TBP recruitment, or overall Pol II transcription, but plays a role in mESC growth and self-renewal. Our data further suggest that yeast and mammalian SAGA complexes contribute to transcription regulation by distinct mechanisms.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
SAGA subunits Spt3 and Spt8 act directly and non-redundantly with TFIID in TBP recruitment in the Gcn4 transcriptome.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf598. doi: 10.1093/nar/gkaf598. Nucleic Acids Res. 2025. PMID: 40637224 Free PMC article.
-
The related coactivator complexes SAGA and ATAC control embryonic stem cell self-renewal through acetyltransferase-independent mechanisms.Cell Rep. 2021 Aug 24;36(8):109598. doi: 10.1016/j.celrep.2021.109598. Cell Rep. 2021. PMID: 34433046 Free PMC article.
-
Structure of the transcription coactivator SAGA.Nature. 2020 Jan;577(7792):717-720. doi: 10.1038/s41586-020-1933-5. Epub 2020 Jan 22. Nature. 2020. PMID: 31969703 Free PMC article.
-
Multifaceted activities of the plant SAGA complex.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194613. doi: 10.1016/j.bbagrm.2020.194613. Epub 2020 Jul 31. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32745625 Review.
-
Architecture of the multi-functional SAGA complex and the molecular mechanism of holding TBP.FEBS J. 2021 May;288(10):3135-3147. doi: 10.1111/febs.15563. Epub 2020 Sep 29. FEBS J. 2021. PMID: 32946670 Review.
Cited by
-
An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters.Genes Dev. 2022 Sep 1;36(17-18):985-1001. doi: 10.1101/gad.350026.122. Epub 2022 Oct 27. Genes Dev. 2022. PMID: 36302553 Free PMC article.
-
Significant Prognostic Factor at Age Cut-off of 73 Years for Advanced Ovarian Serous Cystadenocarcinoma Patients: Insights from Real-World Study.Int J Womens Health. 2024 Feb 3;16:203-218. doi: 10.2147/IJWH.S439335. eCollection 2024. Int J Womens Health. 2024. PMID: 38332982 Free PMC article.
References
-
- Helmlinger D., Papai G., Devys D., Tora L.. What do the structures of GCN5-containing complexes teach us about their function?. Biochim. Biophys. Acta Gene Regul. Mech. 2021; 1864:194614. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

