The DNA methylation landscape of the root-knot nematode-induced pseudo-organ, the gall, in Arabidopsis, is dynamic, contrasting over time, and critically important for successful parasitism
- PMID: 35872574
- PMCID: PMC9825882
- DOI: 10.1111/nph.18395
The DNA methylation landscape of the root-knot nematode-induced pseudo-organ, the gall, in Arabidopsis, is dynamic, contrasting over time, and critically important for successful parasitism
Abstract
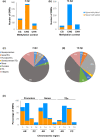
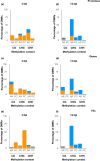
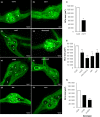
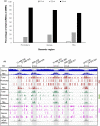
Root-knot nematodes (RKNs) induce giant cells (GCs) within galls which are characterized by large-scale gene repression at early stages. However, the epigenetic mechanism(s) underlying gene silencing is (are) still poorly characterized. DNA methylation in Arabidopsis galls induced by Meloidogyne javanica was studied at crucial infection stages (3 d post-infection (dpi) and 14 dpi) using enzymatic, cytological, and sequencing approaches. DNA methyltransferase mutants (met1, cmt2, cmt3, cmt2/3, drm1/2, ddc) and a DNA demethylase mutant (ros1), were analyzed for RKN resistance/tolerance, and galls were characterized by confocal microscopy and RNA-seq. Early galls were hypermethylated, and the GCs were found to be the major contributors to this hypermethylation, consistent with the very high degree of gene repression they exhibit. By contrast, medium/late galls showed no global increase in DNA methylation compared to uninfected roots, but exhibited large-scale redistribution of differentially methylated regions (DMRs). In line with these findings, it was also shown that DNA methylation and demethylation mutants showed impaired nematode reproduction and gall/GC-development. Moreover, siRNAs that were exclusively present in early galls accumulated at hypermethylated DMRs, overlapping mostly with retrotransposons in the CHG/CG contexts that might be involved in their repression, contributing to their stability/genome integrity. Promoter/gene methylation correlated with differentially expressed genes encoding proteins with basic cell functions. Both mechanisms are consistent with reprogramming host tissues for gall/GC formation. In conclusion, RNA-directed DNA methylation (RdDM; DRM2/1) pathways, maintenance methyltransferases (MET1/CMT3) and demethylation (ROS1) appear to be prominent mechanisms driving a dynamic regulation of the epigenetic landscape during RKN infection.
Keywords: Meloidogyne javanica; Arabidopsis; DNA methylation/epigenetics signatures; galls; giant cells; siRNAs; tomato; transposons.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures




References
-
- de Almeida Engler EJ, Gheysen G. 2013. Nematode‐induced endoreduplication in plant host cells: why and how? Molecular Plant–Microbe Interactions 26: 17–24. - PubMed
-
- de Almeida Engler J, Vieira P, Rodiuc N, Grossi de Sa MF, Engler G. 2015. The plant cell cycle machinery: usurped and modulated by plant‐parasitic nematodes. Advances in Botanical Research 73: 91–118.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

