Molecular Basis of the Slow Growth of Mycoplasma hominis on Different Energy Sources
- PMID: 35873139
- PMCID: PMC9301678
- DOI: 10.3389/fcimb.2022.918557
Molecular Basis of the Slow Growth of Mycoplasma hominis on Different Energy Sources
Abstract
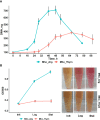
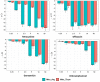
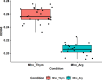
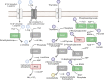
Mycoplasma hominis is an opportunistic urogenital pathogen in vertebrates. It is a non-glycolytic species that produces energy via arginine degradation. Among genital mycoplasmas, M. hominis is the most commonly reported to play a role in systemic infections and can persist in the host for a long time. However, it is unclear how M. hominis proceeds under arginine limitation. The recent metabolic reconstruction of M. hominis has demonstrated its ability to catabolize deoxyribose phosphate to produce ATP. In this study, we cultivated M. hominis on two different energy sources (arginine and thymidine) and demonstrated the differences in growth rate, antibiotic sensitivity, and biofilm formation. Using label-free quantitative proteomics, we compared the proteome of M. hominis under these conditions. A total of 466 proteins were identified from M. hominis, representing approximately 85% of the predicted proteome, while the levels of 94 proteins changed significantly. As expected, we observed changes in the levels of metabolic enzymes. The energy source strongly affects the synthesis of enzymes related to RNA modifications and ribosome assembly. The translocation of lipoproteins and other membrane-associated proteins was also impaired. Our study, the first global characterization of the proteomic switching of M. hominis in arginine-deficiency media, illustrates energy source-dependent control of pathogenicity factors and can help to determine the mechanisms underlying the interaction between the growth rate and fitness of genome-reduced bacteria.
Keywords: Mycoplasma hominis; antibiotic sensitivity; proteomics; slow growth; thymidine.
Copyright © 2022 Evsyutina, Semashko, Galyamina, Kovalchuk, Ziganshin, Ladygina, Fisunov and Pobeguts.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Thymidine utilisation pathway is a novel phenotypic switch of Mycoplasma hominis.J Med Microbiol. 2022 Jan;71(1):001468. doi: 10.1099/jmm.0.001468. J Med Microbiol. 2022. PMID: 35037614 Free PMC article.
-
Comparative Proteomic Analysis of Mycoplasma hominis Grown on Media with Different Carbon Sources.Bull Exp Biol Med. 2021 Aug;171(4):449-452. doi: 10.1007/s10517-021-05247-8. Epub 2021 Sep 20. Bull Exp Biol Med. 2021. PMID: 34542749
-
Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas.PLoS Genet. 2009 Oct;5(10):e1000677. doi: 10.1371/journal.pgen.1000677. Epub 2009 Oct 9. PLoS Genet. 2009. PMID: 19816563 Free PMC article.
-
Molecular dissection of Mycoplasma hominis.APMIS Suppl. 2000;97:1-45. APMIS Suppl. 2000. PMID: 10721331 Review.
-
Mycoplasma hominis: An under recognized pathogen.Indian J Med Microbiol. 2021 Jan;39(1):88-97. doi: 10.1016/j.ijmmb.2020.10.020. Epub 2020 Dec 11. Indian J Med Microbiol. 2021. PMID: 33610259 Review.
Cited by
-
Fluoroquinolones and Biofilm: A Narrative Review.Pharmaceuticals (Basel). 2024 Dec 11;17(12):1673. doi: 10.3390/ph17121673. Pharmaceuticals (Basel). 2024. PMID: 39770514 Free PMC article. Review.
-
Chronic osteomyelitis with pathological fracture induced by Mycoplasma hominis infection: a case report and review of the literature.Front Med (Lausanne). 2025 Jan 27;12:1510753. doi: 10.3389/fmed.2025.1510753. eCollection 2025. Front Med (Lausanne). 2025. PMID: 39931564 Free PMC article.
-
Unraveling the adaptive strategies of Mycoplasma hominis through proteogenomic profiling of clinical isolates.Front Cell Infect Microbiol. 2024 May 2;14:1398706. doi: 10.3389/fcimb.2024.1398706. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38756231 Free PMC article.
-
Changes in vaginal Ureaplasma and Lactobacillus due to antibiotic regimen for premature rupture of membranes.PLoS One. 2025 Feb 18;20(2):e0306958. doi: 10.1371/journal.pone.0306958. eCollection 2025. PLoS One. 2025. PMID: 39964988 Free PMC article.
-
A genome-wide investigation of Mycoplasma hominis genes associated with gynecological infections or infertility.Front Microbiol. 2025 Apr 30;16:1561378. doi: 10.3389/fmicb.2025.1561378. eCollection 2025. Front Microbiol. 2025. PMID: 40371111 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

