Identification and Functional Prediction of Poplar Root circRNAs Involved in Treatment With Different Forms of Nitrogen
- PMID: 35874008
- PMCID: PMC9305699
- DOI: 10.3389/fpls.2022.941380
Identification and Functional Prediction of Poplar Root circRNAs Involved in Treatment With Different Forms of Nitrogen
Abstract
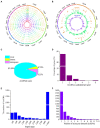
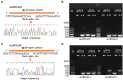
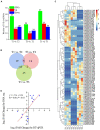
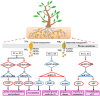
Circular RNAs (circRNAs) are a class of noncoding RNA molecules with ring structures formed by covalent bonds and are commonly present in organisms, playing an important regulatory role in plant growth and development. However, the mechanism of circRNAs in poplar root responses to different forms of nitrogen (N) is still unclear. In this study, high-throughput sequencing was used to identify and predict the function of circRNAs in the roots of poplar exposed to three N forms [1 mM NO3 - (T1), 0.5 mM NH4NO3 (T2, control) and 1 mM NH4 + (T3)]. A total of 2,193 circRNAs were identified, and 37, 24 and 45 differentially expressed circRNAs (DECs) were screened in the T1-T2, T3-T2 and T1-T3 comparisons, respectively. In addition, 30 DECs could act as miRNA sponges, and several of them could bind miRNA family members that play key roles in response to different N forms, indicating their important functions in response to N and plant growth and development. Furthermore, we generated a competing endogenous RNA (ceRNA) regulatory network in poplar roots treated with three N forms. DECs could participate in responses to N in poplar roots through the ceRNA regulatory network, which mainly included N metabolism, amino acid metabolism and synthesis, response to NO3 - or NH4 + and remobilization of N. Together, these results provide new insights into the potential role of circRNAs in poplar root responses to different N forms.
Keywords: Populus × canescens; ceRNA regulatory network; circular RNAs; different nitrogen forms; roots.
Copyright © 2022 Zhou, Yang, Jia, Shi, Deng and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bi Y. M., Zhang Y., Signorelli T., Zhao R., Zhu T., Rothstein S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44, 680–692. doi: 10.1111/j.1365-313X.2005.02568.x, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

