Exchange Broadening Underlies the Enhancement of IDE-Dependent Degradation of Insulin by Anionic Membranes
- PMID: 35874268
- PMCID: PMC9301717
- DOI: 10.1021/acsomega.2c02747
Exchange Broadening Underlies the Enhancement of IDE-Dependent Degradation of Insulin by Anionic Membranes
Abstract
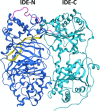
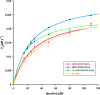
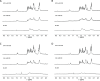
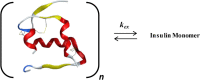
Insulin-degrading enzyme (IDE) is an evolutionarily conserved ubiquitous zinc metalloprotease implicated in the efficient degradation of insulin monomer. However, IDE also degrades monomers of amyloidogenic peptides associated with disease, complicating the development of IDE inhibitors. In this work, we investigated the effects of the lipid composition of membranes on the IDE-dependent degradation of insulin. Kinetic analysis based on chromatography and insulin's helical circular dichroic signal showed that the presence of anionic lipids in membranes enhances IDE's activity toward insulin. Using NMR spectroscopy, we discovered that exchange broadening underlies the enhancement of IDE's activity. These findings, together with the adverse effects of anionic membranes in the self-assembly of IDE's amyloidogenic substrates, suggest that the lipid composition of membranes is a key determinant of IDE's ability to balance the levels of its physiologically and pathologically relevant substrates and achieve proteostasis.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Modulation of the Activity of the Insulin-Degrading Enzyme by Aβ Peptides.ACS Chem Neurosci. 2023 Aug 16;14(16):2935-2943. doi: 10.1021/acschemneuro.3c00384. Epub 2023 Jul 27. ACS Chem Neurosci. 2023. PMID: 37498802
-
Differential Effects of Polyphenols on Insulin Proteolysis by the Insulin-Degrading Enzyme.Antioxidants (Basel). 2021 Aug 25;10(9):1342. doi: 10.3390/antiox10091342. Antioxidants (Basel). 2021. PMID: 34572974 Free PMC article.
-
Enzyme kinetics from circular dichroism of insulin reveals mechanistic insights into the regulation of insulin-degrading enzyme.Biosci Rep. 2018 Nov 7;38(6):BSR20181416. doi: 10.1042/BSR20181416. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30305381 Free PMC article.
-
Modulation of Insulin Sensitivity by Insulin-Degrading Enzyme.Biomedicines. 2021 Jan 17;9(1):86. doi: 10.3390/biomedicines9010086. Biomedicines. 2021. PMID: 33477364 Free PMC article. Review.
-
Insulin-Degrading Enzyme, an Under-Estimated Potential Target to Treat Cancer?Cells. 2022 Apr 5;11(7):1228. doi: 10.3390/cells11071228. Cells. 2022. PMID: 35406791 Free PMC article. Review.
References
-
- Milardi D.; Gazit E.; Radford S. E.; Xu Y.; Gallardo R. U.; Caflisch A.; Westermark G. T.; Westermark P.; Rosa C.; Ramamoorthy A. Proteostasis of islet amyloid polypeptide: A molecular perspective of risk factors and protective strategies for type II diabetes. Chem. Rev. 2021, 121, 1845–1893. 10.1021/acs.chemrev.0c00981. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

