Implementing a Scoring Function Based on Interaction Fingerprint for Autogrow4: Protein Kinase CK1δ as a Case Study
- PMID: 35874609
- PMCID: PMC9301033
- DOI: 10.3389/fmolb.2022.909499
Implementing a Scoring Function Based on Interaction Fingerprint for Autogrow4: Protein Kinase CK1δ as a Case Study
Abstract
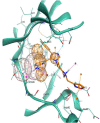
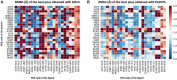
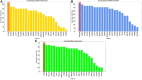
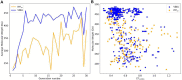
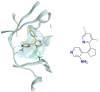
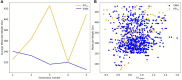
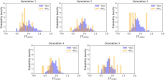
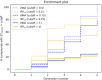
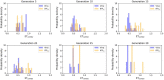
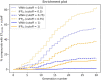
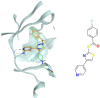
In the last 20 years, fragment-based drug discovery (FBDD) has become a popular and consolidated approach within the drug discovery pipeline, due to its ability to bring several drug candidates to clinical trials, some of them even being approved and introduced to the market. A class of targets that have proven to be particularly suitable for this method is represented by kinases, as demonstrated by the approval of BRAF inhibitor vemurafenib. Within this wide and diverse set of proteins, protein kinase CK1δ is a particularly interesting target for the treatment of several widespread neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis. Computational methodologies, such as molecular docking, are already routinely and successfully applied in FBDD campaigns alongside experimental techniques, both in the hit-discovery and in the hit-optimization stage. Concerning this, the open-source software Autogrow, developed by the Durrant lab, is a semi-automated computational protocol that exploits a combination between a genetic algorithm and a molecular docking software for de novo drug design and lead optimization. In the current work, we present and discuss a modified version of the Autogrow code that implements a custom scoring function based on the similarity between the interaction fingerprint of investigated compounds and a crystal reference. To validate its performance, we performed both a de novo and a lead-optimization run (as described in the original publication), evaluating the ability of our fingerprint-based protocol to generate compounds similar to known CK1δ inhibitors based on both the predicted binding mode and the electrostatic and shape similarity in comparison with the standard Autogrow protocol.
Keywords: Autogrow; de novo drug design; fragment growing; fragment-based drug discovery; interaction fingerprint; lead optimization; neurodegenerative diseases; protein kinase CK1δ.
Copyright © 2022 Pavan, Menin, Bassani, Sturlese and Moro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- AutoGrow4 (2020). AutoGrow4 – Durrant Lab. Available at: https://durrantlab.pitt.edu/autogrow4/ .
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

