SerpinB3 as a Pro-Inflammatory Mediator in the Progression of Experimental Non-Alcoholic Fatty Liver Disease
- PMID: 35874657
- PMCID: PMC9304805
- DOI: 10.3389/fimmu.2022.910526
SerpinB3 as a Pro-Inflammatory Mediator in the Progression of Experimental Non-Alcoholic Fatty Liver Disease
Abstract
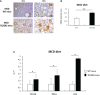
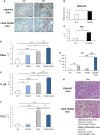
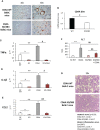
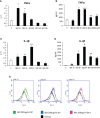
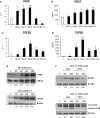
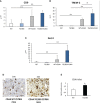
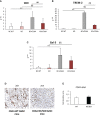
Non-alcoholic fatty liver disease (NAFLD) is becoming the most common chronic liver disease worldwide. In 20-30% of patients, NAFLD can progress into non-alcoholic steatohepatitis (NASH), eventually leading to fibrosis, cirrhosis and hepatocellular carcinoma development. SerpinB3 (SB3), a hypoxia-inducible factor-2α dependent cysteine protease inhibitor, is up-regulated in hepatocytes during progressive NAFLD and proposed to contribute to disease progression. In this study we investigated the pro-inflammatory role of SB3 by employing phorbol-myristate acetate-differentiated human THP-1 macrophages exposed in vitro to human recombinant SB3 (hrSB3) along with mice overexpressing SB3 in hepatocytes (TG/SB3) or knockout for SB3 (KO/SB3) in which NASH was induced by feeding methionine/choline deficient (MCD) or a choline-deficient, L-amino acid defined (CDAA) diets. In vivo experiments showed that the induction of NASH in TG/SB3 mice was characterized by an impressive increase of liver infiltrating macrophages that formed crown-like aggregates and by an up-regulation of hepatic transcript levels of pro-inflammatory cytokines. All these parameters and the extent of liver damage were significantly blunted in KO/SB3 mice. In vitro experiments confirmed that hrSB3 stimulated macrophage production of M1-cytokines such as TNFα and IL-1β and reactive oxygen species along with that of TGFβ and VEGF through the activation of the NF-kB transcription factor. The opposite changes in liver macrophage activation observed in TG/SB3 or KO/SB3 mice with NASH were associated with a parallel modulation in the expression of triggering receptor expressed on myeloid cells-2 (TREM2), CD9 and galectin-3 markers, recently detected in NASH-associated macrophages. From these results we propose that SB3, produced by activated/injured hepatocytes, may operate as a pro-inflammatory mediator in NASH contributing to the disease progression.
Keywords: NASH; SerpinB3; hepatocytes; innate immunity; macrophages.
Copyright © 2022 Novo, Cappon, Villano, Quarta, Cannito, Bocca, Turato, Guido, Maggiora, Protopapa, Sutti, Provera, Ruvoletto, Biasiolo, Foglia, Albano, Pontisso and Parola.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

