Added Value of Reanalysis of Whole Exome- and Whole Genome Sequencing Data From Patients Suspected of Primary Immune Deficiency Using an Extended Gene Panel and Structural Variation Calling
- PMID: 35874679
- PMCID: PMC9302041
- DOI: 10.3389/fimmu.2022.906328
Added Value of Reanalysis of Whole Exome- and Whole Genome Sequencing Data From Patients Suspected of Primary Immune Deficiency Using an Extended Gene Panel and Structural Variation Calling
Abstract
Background: Knowledge of the genetic variation underlying Primary Immune Deficiency (PID) is increasing. Reanalysis of genome-wide sequencing data from undiagnosed patients with suspected PID may improve the diagnostic rate.
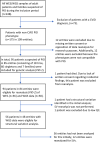
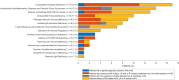
Methods: We included patients monitored at the Department of Infectious Diseases or the Child and Adolescent Department, Rigshospitalet, Denmark, for a suspected PID, who had been analysed previously using a targeted PID gene panel (457 PID-related genes) on whole exome- (WES) or whole genome sequencing (WGS) data. A literature review was performed to extend the PID gene panel used for reanalysis of single nucleotide variation (SNV) and small indels. Structural variant (SV) calling was added on WGS data.
Results: Genetic data from 94 patients (86 adults) including 36 WES and 58 WGS was reanalysed a median of 23 months after the initial analysis. The extended gene panel included 208 additional PID-related genes. Genetic reanalysis led to a small increase in the proportion of patients with new suspicious PID related variants of uncertain significance (VUS). The proportion of patients with a causal genetic diagnosis was constant. In total, five patients (5%, including three WES and two WGS) had a new suspicious PID VUS identified due to reanalysis. Among these, two patients had a variant added due to the expansion of the PID gene panel, and three patients had a variant reclassified to a VUS in a gene included in the initial PID gene panel. The total proportion of patients with PID related VUS, likely pathogenic, and pathogenic variants increased from 43 (46%) to 47 (50%), as one patient had a VUS detected in both initial- and reanalysis. In addition, we detected new suspicious SNVs and SVs of uncertain significance in PID candidate genes with unknown inheritance and/or as heterozygous variants in genes with autosomal recessive inheritance in 8 patients.
Conclusion: These data indicate a possible diagnostic gain of reassessing WES/WGS data from patients with suspected PID. Reasons for the possible gain included improved knowledge of genotype-phenotype correlation, expanding the gene panel, and adding SV analyses. Future studies of genotype-phenotype correlations may provide additional knowledge on the impact of the new suspicious VUSs.
Keywords: gene panel analysis; primary immune deficiencies (PID); reanalysis approach; single nucleotide variant analysis; small INDELs; structural variation analysis; whole exome sequencing; whole genome sequencing (WGS).
Copyright © 2022 Mørup, Nazaryan-Petersen, Gabrielaite, Reekie, Marquart, Hartling, Marvig, Katzenstein, Masmas, Lundgren, Murray, Helleberg and Borgwardt.
Conflict of interest statement
TK has participated in advisory boards for Gilead Sciences, GlaxoSmithKline/ViiV and MSD. TK has received fees for teaching from Takeda and CSL Behring, and a research grant from Gilead Sciences. MH has participated in advisory boards for AstraZeneca, Gilead Sciences, GlaxoSmithKline/ViiV, MSD, Roche, and Sobi. MH has received fees for teaching from Gilead Sciences and GSK. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. Human Inborn Errors of Immunity: 2019 Update on the Classification From the International Union of Immunological Societies Expert Committee. J Clin Immunol (2020) 40:24–64. doi: 10.1007/s10875-019-00737-x - DOI - PMC - PubMed
-
- Yska HAF, Elsink K, Kuijpers TW, Frederix GWJ, van Gijn ME, van Montfrans JM. Diagnostic Yield of Next Generation Sequencing in Genetically Undiagnosed Patients With Primary Immunodeficiencies: A Systematic Review. J Clin Immunol (2019) 39:577–91. doi: 10.1007/s10875-019-00656-x - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

