Single-cell transcriptomes underscore genetically distinct tumor characteristics and microenvironment for hereditary kidney cancers
- PMID: 35874919
- PMCID: PMC9301876
- DOI: 10.1016/j.isci.2022.104463
Single-cell transcriptomes underscore genetically distinct tumor characteristics and microenvironment for hereditary kidney cancers
Abstract
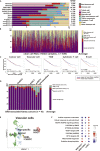
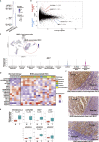
Our understanding of how each hereditary kidney cancer adapts to its tissue microenvironment is incomplete. Here, we present single-cell transcriptomes of 108,342 cells from patient specimens including from six hereditary kidney cancers. The transcriptomes displayed distinct characteristics of the cell of origin and unique tissue microenvironment for each hereditary kidney cancer. Of note, hereditary leiomyomatosis and renal cell carcinoma (HLRCC)-associated kidney cancer retained some characteristics of proximal tubules, which were completely lost in lymph node metastases and present as an avascular tumor with suppressed T cells and TREM2-high macrophages, leading to immune tolerance. Birt-Hogg-Dubé (BHD)-associated kidney cancer exhibited transcriptomic intratumor heterogeneity (tITH) with increased characteristics of intercalated cells of the collecting duct and upregulation of FOXI1-driven genes, a critical transcription factor for collecting duct differentiation. These findings facilitate our understanding of how hereditary kidney cancers adapt to their tissue microenvironment.
Keywords: Cancer; Cancer systems biology; Human specimen; Microenvironment; Oncology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Aird W.C. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ. Res. 2007;100:174–190. doi: 10.1161/01.res.0000255690.03436.ae. - DOI - PubMed
-
- Ball M.W., An J.Y., Gomella P.T., Gautam R., Ricketts C.J., Vocke C.D., Schmidt L.S., Merino M.J., Srinivasan R., Malayeri A.A., et al. Growth rates of genetically defined renal tumors: implications for active surveillance and intervention. J. Clin. Oncol. 2020;38:1146–1153. doi: 10.1200/jco.19.02263. - DOI - PMC - PubMed
-
- Barone S., Zahedi K., Brooks M., Henske E.P., Yang Y., Zhang E., Bissler J.J., Yu J.J., Soleimani M. Kidney intercalated cells and the transcription factor FOXi1 drive cystogenesis in tuberous sclerosis complex. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2020190118. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

