ContactPFP: Protein function prediction using predicted contact information
- PMID: 35875419
- PMCID: PMC9302406
- DOI: 10.3389/fbinf.2022.896295
ContactPFP: Protein function prediction using predicted contact information
Abstract
Computational function prediction is one of the most important problems in bioinformatics as elucidating the function of genes is a central task in molecular biology and genomics. Most of the existing function prediction methods use protein sequences as the primary source of input information because the sequence is the most available information for query proteins. There are attempts to consider other attributes of query proteins. Among these attributes, the three-dimensional (3D) structure of proteins is known to be very useful in identifying the evolutionary relationship of proteins, from which functional similarity can be inferred. Here, we report a novel protein function prediction method, ContactPFP, which uses predicted residue-residue contact maps as input structural features of query proteins. Although 3D structure information is known to be useful, it has not been routinely used in function prediction because the 3D structure is not experimentally determined for many proteins. In ContactPFP, we overcome this limitation by using residue-residue contact prediction, which has become increasingly accurate due to rapid development in the protein structure prediction field. ContactPFP takes a query protein sequence as input and uses predicted residue-residue contact as a proxy for the 3D protein structure. To characterize how predicted contacts contribute to function prediction accuracy, we compared the performance of ContactPFP with several well-established sequence-based function prediction methods. The comparative study revealed the advantages and weaknesses of ContactPFP compared to contemporary sequence-based methods. There were many cases where it showed higher prediction accuracy. We examined factors that affected the accuracy of ContactPFP using several illustrative cases that highlight the strength of our method.
Keywords: PFP; contact prediction; function annotation; function prediction; functional genomics; gene function; protein structure; residue contacts.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


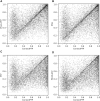





Similar articles
-
Improving AlphaFold Predicted Contacts for Alpha-Helical Transmembrane Proteins Using Structural Features.Int J Mol Sci. 2024 May 11;25(10):5247. doi: 10.3390/ijms25105247. Int J Mol Sci. 2024. PMID: 38791287 Free PMC article.
-
Identification of residue pairing in interacting β-strands from a predicted residue contact map.BMC Bioinformatics. 2018 Apr 19;19(1):146. doi: 10.1186/s12859-018-2150-1. BMC Bioinformatics. 2018. PMID: 29673311 Free PMC article.
-
Ab initio and template-based prediction of multi-class distance maps by two-dimensional recursive neural networks.BMC Struct Biol. 2009 Jan 30;9:5. doi: 10.1186/1472-6807-9-5. BMC Struct Biol. 2009. PMID: 19183478 Free PMC article.
-
Prediction of protein function from protein sequence and structure.Q Rev Biophys. 2003 Aug;36(3):307-40. doi: 10.1017/s0033583503003901. Q Rev Biophys. 2003. PMID: 15029827 Review.
-
Exploring plant protein functions through structure-based clustering.Trends Plant Sci. 2025 Apr 15:S1360-1385(25)00091-3. doi: 10.1016/j.tplants.2025.03.014. Online ahead of print. Trends Plant Sci. 2025. PMID: 40240260 Review.
Cited by
-
TEMPROT: protein function annotation using transformers embeddings and homology search.BMC Bioinformatics. 2023 Jun 8;24(1):242. doi: 10.1186/s12859-023-05375-0. BMC Bioinformatics. 2023. PMID: 37291492 Free PMC article.
-
Lipid Trafficking in Diverse Bacteria.Acc Chem Res. 2025 Jan 7;58(1):36-46. doi: 10.1021/acs.accounts.4c00540. Epub 2024 Dec 16. Acc Chem Res. 2025. PMID: 39680024 Free PMC article. Review.
-
Domain-PFP allows protein function prediction using function-aware domain embedding representations.Commun Biol. 2023 Oct 31;6(1):1103. doi: 10.1038/s42003-023-05476-9. Commun Biol. 2023. PMID: 37907681 Free PMC article.
-
A machine learning model for the proteome-wide prediction of lipid-interacting proteins.bioRxiv [Preprint]. 2025 May 25:2024.01.26.577452. doi: 10.1101/2024.01.26.577452. bioRxiv. 2025. PMID: 38352308 Free PMC article. Preprint.
-
Domain-PFP: Protein Function Prediction Using Function-Aware Domain Embedding Representations.bioRxiv [Preprint]. 2023 Aug 24:2023.08.23.554486. doi: 10.1101/2023.08.23.554486. bioRxiv. 2023. Update in: Commun Biol. 2023 Oct 31;6(1):1103. doi: 10.1038/s42003-023-05476-9. PMID: 37662252 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

