Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing
- PMID: 35875870
- PMCID: PMC10024945
- DOI: 10.1021/jacs.2c05349
Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing
Abstract
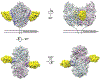
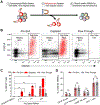
The clinical manufacturing of chimeric antigen receptor (CAR) T cells includes cell selection, activation, gene transduction, and expansion. While the method of T-cell selection varies across companies, current methods do not actively eliminate the cancer cells in the patient's apheresis product from the healthy immune cells. Alarmingly, it has been found that transduction of a single leukemic B cell with the CAR gene can confer resistance to CAR T-cell therapy and lead to treatment failure. In this study, we report the identification of a novel high-affinity DNA aptamer, termed tJBA8.1, that binds transferrin receptor 1 (TfR1), a receptor broadly upregulated by cancer cells. Using competition assays, high resolution cryo-EM, and de novo model building of the aptamer into the resulting electron density, we reveal that tJBA8.1 shares a binding site on TfR1 with holo-transferrin, the natural ligand of TfR1. We use tJBA8.1 to effectively deplete B lymphoma cells spiked into peripheral blood mononuclear cells with minimal impact on the healthy immune cell composition. Lastly, we present opportunities for affinity improvement of tJBA8.1. As TfR1 expression is broadly upregulated in many cancers, including difficult-to-treat T-cell leukemias and lymphomas, our work provides a facile, universal, and inexpensive approach for comprehensively removing cancerous cells from patient apheresis products for safe manufacturing of adoptive T-cell therapies.
Conflict of interest statement
The authors declare the following competing financial interest(s): Michael Jensen has interests in Umoja Biopharma and Juno Therapeutics, a Bristol-Myers Squibb company. Michael Jensen is a seed investor and holds ownership equity in Umoja, serves as a member of the Umoja Joint Steering Committee, and is a Board Observer of the Umoja Board of Directors. Michael Jensen holds patents, some of which are licensed to Umoja Biopharma and Juno Therapeutics. Suzie Pun, Nataly Kacherovsky, Emmeline Cheng, Ian Cardle, and Michael Jensen are co-inventors on a provisional patent application for the aptamers and cell depletion strategy described in this article.
Figures




References
-
- Maude SL; Laetsch TW; Buechner J; Rives S; Boyer M; Bittencourt H; Bader P; Verneris MR; Stefanski HE; Myers GD; Qayed M; De Moerloose B; Hiramatsu H; Schlis K; Davis KL; Martin PL; Nemecek ER; Yanik GA; Peters C; Baruchel A; Boissel N; Mechinaud F; Balduzzi A; Krueger J; June CH; Levine BL; Wood P; Taran T; Leung M; Mueller KT; Zhang Y; Sen K; Lebwohl D; Pulsipher MA; Grupp SA Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. New England Journal of Medicine 2018, 378 (5), 439–448. - PMC - PubMed
-
- Schuster SJ; Bishop MR; Tam CS; Waller EK; Borchmann P; McGuirk JP; Jäger U; Jaglowski S; Andreadis C; Westin JR; Fleury I; Bachanova V; Foley SR; Ho PJ; Mielke S; Magenau JM; Holte H; Pantano S; Pacaud LB; Awasthi R; Chu J; Anak Ö; Salles G; Maziarz RT Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. New England Journal of Medicine 2018, 380 (1), 45–56. - PubMed
-
- Neelapu SS; Locke FL; Bartlett NL; Lekakis LJ; Miklos DB; Jacobson CA; Braunschweig I; Oluwole OO; Siddiqi T; Lin Y; Timmerman JM; Stiff PJ; Friedberg JW; Flinn IW; Goy A; Hill BT; Smith MR; Deol A; Farooq U; McSweeney P; Munoz J; Avivi I; Castro JE; Westin JR; Chavez JC; Ghobadi A; Komanduri KV; Levy R; Jacobsen ED; Witzig TE; Reagan P; Bot A; Rossi J; Navale L; Jiang Y; Aycock J; Elias M; Chang D; Wiezorek J; Go WY Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. New England Journal of Medicine 2017, 377 (26), 2531–2544. - PMC - PubMed
-
- Locke FL; Ghobadi A; Jacobson CA; Miklos DB; Lekakis LJ; Oluwole OO; Lin Y; Braunschweig I; Hill BT; Timmerman JM; Deol A; Reagan PM; Stiff P; Flinn IW; Farooq U; Goy A; McSweeney PA; Munoz J; Siddiqi T; Chavez JC; Herrera AF; Bartlett NL; Wiezorek JS; Navale L; Xue A; Jiang Y; Bot A; Rossi JM; Kim JJ; Go WY; Neelapu SS Long-Term Safety and Activity of Axicabtagene Ciloleucel in Refractory Large B-Cell Lymphoma (ZUMA-1): A Single-Arm, Multicentre, Phase 1–2 Trial. The Lancet Oncology 2019, 20 (1), 31–42. - PMC - PubMed
-
- Wang M; Munoz J; Goy A; Locke FL; Jacobson CA; Hill BT; Timmerman JM; Holmes H; Jaglowski S; Flinn IW; McSweeney PA; Miklos DB; Pagel JM; Kersten M-J; Milpied N; Fung H; Topp MS; Houot R; Beitinjaneh A; Peng W; Zheng L; Rossi JM; Jain RK; Rao AV; Reagan PM KTE-X19 CAR T-Cell Therapy in Relapsed or Refractory Mantle-Cell Lymphoma. New England Journal of Medicine 2020, 382 (14), 1331–1342. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

