Mutation spectrum in a cohort with familial exudative vitreoretinopathy
- PMID: 35876299
- PMCID: PMC9482396
- DOI: 10.1002/mgg3.2021
Mutation spectrum in a cohort with familial exudative vitreoretinopathy
Abstract
Purpose: To expand the mutation spectrum of patients with familial exudative vitreoretinopathy (FEVR) disease.
Participants: 74 probands (53 families and 21 sporadic probands) with familial exudative vitreoretinopathy (FEVR) disease and their available family members (n = 188) were recruited for sequencing.
Methods: Panel-based targeted screening was performed on all subjects. Before sanger sequencing, variants of LRP5, NDP, FZD4, TSPAN12, ZNF408, KIF11, RCBTB1, JAG1, and CTNNA1 genes were verified by a series of bioinformatics tools and genotype-phenotype co-segregation analysis.
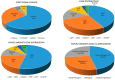
Results: 40.54% (30/74) of the probands were sighted to possess at least one etiological mutation of the nine FEVR-causative genes. The etiological mutation detection rate was 37.74% (20/53) in family-attainable probands while 47.62% (10/21) in sporadic cases. The diagnosis rate of patients in the early-onset subgroup (≤5 years old, 45.4%) is higher than that of the children or adolescence-onset subgroup (6-16 years old, 42.1%) and the late-onset subgroup (≥17 years old, 39.4%). A total of 36 etiological mutations were identified in this study, comprising 26 novel mutations and 10 reported mutations. LRP5 was the most prevalent mutant gene among the 36 mutation types with a percentage of 41.67% (15/36). Followed by FZD4 (10/36, 27.78%), TSPAN12 (5/36, 13.89%), NDP (4/36, 11.11%), KIF11 (1/36, 2.78%), and RCBTB1 (1/36, 2.78%). Among these mutations, 63.89% (23/36) were missense mutations, 25.00% (9/36) were frameshift mutations, 5.56% (2/36) were splicing mutations, 5.56% (2/36) were nonsense mutations. Moreover, the clinical pathogenicity of these variants was defined according to American College of Medical Genetics (ACMG) and genomics guidelines: 41.67% (15/36) were likely pathogenic variants, 27.78% (10/36) pathogenic variants, 30.55% (11/36) variants of uncertain significance. No etiological mutations discovered in the ZNF408, JAG1, and CTNNA1 genes in this FEVR cohort.
Conclusions: We systematically screened nine FEVR disease-associated genes in a cohort of 74 Chinese probands with FEVR disease. With a detection rate of 40.54%, 36 etiological mutations of six genes were authenticated in 30 probands, including 26 novel mutations and 10 reported mutations. The most prevalent mutated gene is LRP5, followed by FZD4, TSPAN12, NDP, KIF11, and RCBTB1. In total, a de novo mutation was confirmed. Our study significantly clarified the mutation spectrum of variants bounded up to FEVR disease.
Keywords: FEVR; genotype-phenotype analysis; mutation Spectrum; targeted sequencing.
© 2022 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Chen, Z. Y. , Battinelli, E. M. , Fielder, A. , Bundey, S. , Sims, K. , Breakefield, X. O. , & Craig, I. W. (1993). A mutation in the Norrie disease gene (NDP) associated with X‐linked familial exudative vitreoretinopathy. Nature Genetics, 5(2), 180–183. - PubMed
-
- Collin, R. W. , Nikopoulos, K. , Dona, M. , Gilissen, C. , Hoischen, A. , Boonstra, F. N. , Poulter, J. A. , Kondo, H. , Berger, W. , Toomes, C. , Tahira, T. , Mohn, L. R. , Blokland, E. A. , Hetterschijt, L. , Ali, M. , Groothuismink, J. M. , Duijkers, L. , Inglehearn, C. F. , Sollfrank, L. , … Cremers, F. P. M. (2013). ZNF408 is mutated in familial exudative vitreoretinopathy and is crucial for the development of zebrafish retinal vasculature. Proceedings of the National Academy of Sciences of the United States of America, 110(24), 9856–9861. - PMC - PubMed
-
- Criswick, V. G. , & Schepens, C. L. (1969). Familial exudative vitreoretinopathy. American Journal of Ophthalmology, 68(4), 578–594. - PubMed
-
- Dixon, M. W. , Stem, M. S. , Schuette, J. L. , Keegan, C. E. , & Besirli, C. G. (2016). CTNNB1 mutation associated with familial exudative vitreoretinopathy (FEVR) phenotype. Ophthalmic Genetics, 37(4), 468–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

