Multiple Mechanisms Confer Resistance to Azithromycin in Shigella in Bangladesh: a Comprehensive Whole Genome-Based Approach
- PMID: 35876510
- PMCID: PMC9430107
- DOI: 10.1128/spectrum.00741-22
Multiple Mechanisms Confer Resistance to Azithromycin in Shigella in Bangladesh: a Comprehensive Whole Genome-Based Approach
Abstract
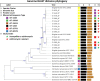
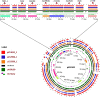
Shigella is the second leading cause of diarrheal deaths worldwide. Azithromycin (AZM) is a potential treatment option for Shigella infection; however, the recent emergence of AZM resistance in Shigella threatens the current treatment strategy. Therefore, we conducted a comprehensive whole genome-based approach to identify the mechanism(s) of AZM resistance in Shigella. We performed antimicrobial susceptibility tests, polymerase chain reaction (PCR), Sanger (amplicon) sequencing, and whole genome-based bioinformatics approaches to conduct the study. Fifty-seven (38%) of the Shigella isolates examined were AZM resistant; Shigella sonnei exhibited the highest rate of resistance against AZM (80%). PCR amplification for 15 macrolide resistance genes (MRGs) followed by whole-genome analysis of 13 representative Shigella isolates identified two AZM-modifying genes, mph(A) (in all Shigella isolates resistant to AZM) and mph(E) (in 2 AZM-resistant Shigella isolates), as well as one 23S rRNA-methylating gene, erm(B) (41% of AZM-resistant Shigella isolates) and one efflux pump mediator gene, msr(E) [in the same two Shigella isolates that harbored the mph(E) gene]. This is the first report of msr(E) and mph(E) genes in Shigella. Moreover, we found that an IncFII-type plasmid predominates and can possess all four MRGs. We also detected two plasmid-borne resistance gene clusters: IS26-mph(A)-mrx(A)-mph(R)(A)-IS6100, which is linked to global dissemination of MRGs, and mph(E)-msr(E)-IS482-IS6, which is reported for the first time in Shigella. In conclusion, this study demonstrates that MRGs in association with pathogenic IS6 family insertion sequences generate resistance gene clusters that propagate through horizontal gene transfer (HGT) in Shigella. IMPORTANCE Shigella can frequently transform into a superbug due to uncontrolled and rogue administration of antibiotics and the emergence of HGT of antimicrobial resistance factors. The advent of AZM resistance in Shigella has become a serious concern in the treatment of shigellosis. However, there is an obvious scarcity of clinical data and research on genetic mechanisms that induce AZM resistance in Shigella, particularly in low- and middle-income countries. Therefore, this study is an approach to raise the alarm for the next lifeline. We show that two key MRGs [mph(A) and erm(B)] and the newly identified MRGs [mph(E) and msr(E)], with their origination in plasmid-borne pathogenic islands, are fundamental mechanisms of AZM resistance in Shigella in Bangladesh. Overall, this study predicts an abrupt decrease in the effectiveness of AZM against Shigella in the very near future and suggests prompt focus on seeking a more effective treatment alternative to AZM for shigellosis.
Keywords: Shigella; antimicrobial resistance mechanism; azithromycin; resistance gene cluster; whole genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Multidrug-resistant conjugative plasmid carrying mphA confers increased antimicrobial resistance in Shigella.Sci Rep. 2024 Mar 23;14(1):6947. doi: 10.1038/s41598-024-57423-1. Sci Rep. 2024. PMID: 38521802 Free PMC article.
-
Resistome phylodynamics of multidrug-resistant Shigella isolated from diarrheal patients.Microbiol Spectr. 2025 Jan 7;13(1):e0163524. doi: 10.1128/spectrum.01635-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39612215 Free PMC article.
-
Evaluation of Shigella Species Azithromycin CLSI Epidemiological Cutoff Values and Macrolide Resistance Genes.J Clin Microbiol. 2019 Mar 28;57(4):e01422-18. doi: 10.1128/JCM.01422-18. Print 2019 Apr. J Clin Microbiol. 2019. PMID: 30700507 Free PMC article.
-
The increasing antimicrobial resistance of Shigella species among Iranian pediatrics: a systematic review and meta-analysis.Pathog Glob Health. 2023 Oct;117(7):611-622. doi: 10.1080/20477724.2023.2179451. Epub 2023 Feb 16. Pathog Glob Health. 2023. PMID: 36794800 Free PMC article.
-
Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence.Paediatr Int Child Health. 2018 Nov;38(sup1):S50-S65. doi: 10.1080/20469047.2017.1409454. Paediatr Int Child Health. 2018. PMID: 29790845 Free PMC article.
Cited by
-
Plasmid-mediated azithromycin resistance in non-typhoidal Salmonella recovered from human infections.J Antimicrob Chemother. 2024 Oct 1;79(10):2688-2697. doi: 10.1093/jac/dkae281. J Antimicrob Chemother. 2024. PMID: 39119898 Free PMC article.
-
Characterization of enterotoxin, antibiotic resistance genes, and antimicrobial susceptibility profiling of Staphylococcus aureus isolated from table eggs: Implications for food safety and public health.Open Vet J. 2025 Mar;15(3):1187-1205. doi: 10.5455/OVJ.2025.v15.i3.11. Epub 2025 Mar 31. Open Vet J. 2025. PMID: 40276192 Free PMC article.
-
Multidrug-resistant conjugative plasmid carrying mphA confers increased antimicrobial resistance in Shigella.Sci Rep. 2024 Mar 23;14(1):6947. doi: 10.1038/s41598-024-57423-1. Sci Rep. 2024. PMID: 38521802 Free PMC article.
-
Whole-Genome Sequencing of an Escherichia coli ST69 Strain Harboring blaCTX-M-27 on a Hybrid Plasmid.Infect Drug Resist. 2024 Feb 1;17:365-375. doi: 10.2147/IDR.S427571. eCollection 2024. Infect Drug Resist. 2024. PMID: 38318209 Free PMC article.
-
Resistome phylodynamics of multidrug-resistant Shigella isolated from diarrheal patients.Microbiol Spectr. 2025 Jan 7;13(1):e0163524. doi: 10.1128/spectrum.01635-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39612215 Free PMC article.
References
-
- Liu Y, Hu L, Pan Y, Cheng J, Zhu Y, Ye Y, Li J. 2012. Prevalence of plasmid-mediated quinolone resistance determinants in association with β-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J Med Microbiol 61:1174–1176. doi:10.1099/jmm.0.042580-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

