Turning coldspots into hotspots: targeted recruitment of axis protein Hop1 stimulates meiotic recombination in Saccharomyces cerevisiae
- PMID: 35876814
- PMCID: PMC9434160
- DOI: 10.1093/genetics/iyac106
Turning coldspots into hotspots: targeted recruitment of axis protein Hop1 stimulates meiotic recombination in Saccharomyces cerevisiae
Abstract
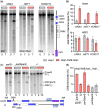
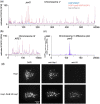
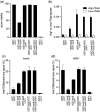
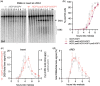
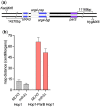
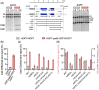
The DNA double-strand breaks that initiate meiotic recombination are formed in the context of the meiotic chromosome axis, which in Saccharomyces cerevisiae contains a meiosis-specific cohesin isoform and the meiosis-specific proteins Hop1 and Red1. Hop1 and Red1 are important for double-strand break formation; double-strand break levels are reduced in their absence and their levels, which vary along the lengths of chromosomes, are positively correlated with double-strand break levels. How axis protein levels influence double-strand break formation and recombination remains unclear. To address this question, we developed a novel approach that uses a bacterial ParB-parS partition system to recruit axis proteins at high levels to inserts at recombination coldspots where Hop1 and Red1 levels are normally low. Recruiting Hop1 markedly increased double-strand breaks and homologous recombination at target loci, to levels equivalent to those observed at endogenous recombination hotspots. This local increase in double-strand breaks did not require Red1 or the meiosis-specific cohesin component Rec8, indicating that, of the axis proteins, Hop1 is sufficient to promote double-strand break formation. However, while most crossovers at endogenous recombination hotspots are formed by the meiosis-specific MutLγ resolvase, crossovers that formed at an insert locus were only modestly reduced in the absence of MutLγ, regardless of whether or not Hop1 was recruited to that locus. Thus, while local Hop1 levels determine local double-strand break levels, the recombination pathways that repair these breaks can be determined by other factors, raising the intriguing possibility that different recombination pathways operate in different parts of the genome.
Keywords: Hop1; chromosome axis; crossing over; double-strand breaks; meiosis; meiotic recombination; recombination pathways.
Published by Oxford University Press on behalf of Genetics Society of America 2022.
Figures



Similar articles
-
Meiotic DNA double-strand break-independent role of protein phosphatase 4 in Hop1 assembly to promote meiotic chromosome axis formation in budding yeast.Genes Cells. 2023 Aug;28(8):595-614. doi: 10.1111/gtc.13049. Epub 2023 May 27. Genes Cells. 2023. PMID: 37243502
-
How do small chromosomes know they are small? Maximizing meiotic break formation on the shortest yeast chromosomes.Curr Genet. 2021 Jun;67(3):431-437. doi: 10.1007/s00294-021-01160-9. Epub 2021 Feb 18. Curr Genet. 2021. PMID: 33604699 Free PMC article. Review.
-
Noncanonical Contributions of MutLγ to VDE-Initiated Crossovers During Saccharomyces cerevisiae Meiosis.G3 (Bethesda). 2019 May 7;9(5):1647-1654. doi: 10.1534/g3.119.400150. G3 (Bethesda). 2019. PMID: 30902890 Free PMC article.
-
Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis.Elife. 2016 Nov 18;5:e19669. doi: 10.7554/eLife.19669. Elife. 2016. PMID: 27855779 Free PMC article.
-
Kinetochores, cohesin, and DNA breaks: Controlling meiotic recombination within pericentromeres.Yeast. 2019 Mar;36(3):121-127. doi: 10.1002/yea.3366. Epub 2019 Feb 3. Yeast. 2019. PMID: 30625250 Free PMC article. Review.
Cited by
-
Exo1 protects DNA nicks from ligation to promote crossover formation during meiosis.PLoS Biol. 2023 Apr 20;21(4):e3002085. doi: 10.1371/journal.pbio.3002085. eCollection 2023 Apr. PLoS Biol. 2023. PMID: 37079643 Free PMC article.
-
Temperature regulates negative supercoils to modulate meiotic crossovers and chromosome organization.Sci China Life Sci. 2024 Nov;67(11):2426-2443. doi: 10.1007/s11427-024-2671-1. Epub 2024 Jul 23. Sci China Life Sci. 2024. PMID: 39048717
-
The ATPase activity of yeast chromosome axis protein Hop1 affects the frequency of meiotic crossovers.Nucleic Acids Res. 2025 Jan 24;53(3):gkae1264. doi: 10.1093/nar/gkae1264. Nucleic Acids Res. 2025. PMID: 39727188 Free PMC article.
-
Absence of chromosome axis protein recruitment prevents meiotic recombination chromosome-wide in the budding yeast Lachancea kluyveri.Proc Natl Acad Sci U S A. 2024 Mar 19;121(12):e2312820121. doi: 10.1073/pnas.2312820121. Epub 2024 Mar 13. Proc Natl Acad Sci U S A. 2024. PMID: 38478689 Free PMC article.
-
Spo11: from topoisomerase VI to meiotic recombination initiator.Biochem Soc Trans. 2025 Apr 2;53(2):383-98. doi: 10.1042/BST20253019. Biochem Soc Trans. 2025. PMID: 40181639 Free PMC article. Review.
References
-
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V.. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339(6116):215–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

